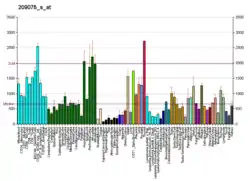
ISCU
Iron-sulfur cluster assembly enzyme ISCU, mitochondrial is a protein that in humans is encoded by the ISCU gene.[5] It encodes an iron-sulfur (Fe-S) cluster scaffold protein involved in [2Fe-2S] and [4Fe-4S] cluster synthesis and maturation.[6][7][8][9] A deficiency of ISCU is associated with a mitochondrial myopathy with lifelong exercise intolerance where only minor exertion causes tachycardia, shortness of breath, muscle weakness and myalgia.[10]
| ISCU | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | ISCU, 2310020H20Rik, HML, ISU2, NIFU, NIFUN, hnifU, iron-sulfur cluster assembly enzyme | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 611911 MGI: 1913633 HomoloGene: 6991 GeneCards: ISCU | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Structure
ISCU is located on the q arm of chromosome 12 in position 23.3 and has 8 exons.[7] ISCU, the protein encoded by this gene, is a member of the NifU family. It is an iron-sulfur transferase that contains binding sites for [2Fe-2S] and [4Fe-4S] clusters. ISCU contains a transit peptide, 4 beta strands, 4 alpha helixes, and 4 turns.[8][9] Alternative splicing results in transcript variants encoding different protein isoforms that localize either to the cytosol or to the mitochondrion. A pseudogene of this gene is present on chromosome 1.[7]
Function
ISCU encodes a component of the iron-sulfur (Fe-S) cluster scaffold responsible for the synthesis and maturation of [2Fe-2S] and [4Fe-4S] clusters. Fe-S clusters are cofactors that play a role in the function of a diverse set of enzymes, including those that regulate metabolism, iron homeostasis, and oxidative stress response. In one process, the [2Fe-2S] cluster transiently assembles on ISCU and is then transferred to GLRX5 in a cysteine desulfurase complex NFS1-LYRM4/ISD11 dependent process.[7][6][8][9]
ISCU has two isoforms, isoform 1, which is found in the mitochondrion and isoform 2, which is found in the nucleus and cytoplasm.[8][9]
Clinical significance
ISCU mutations have been found in patients with hereditary mitochondrial myopathy with exercise intolerance and lactic acidosis. This disease is a result of a deficiency of ISCU that corresponds to the deficiency of mitochondrial iron-sulfur proteins and impaired muscle oxidative metabolism.[7] Characteristics of mitochondrial myopathy with deficiency of ISCU may include lifelong exercise intolerance in which exertion can cause tachycardia, dyspnoea, cardiac palpitations, shortness of breath, fatigue, pain of active muscles, rhabdomyolysis, myoglobinuria, elevated lactate and pyruvate, decreased oxygen utilization, large calves, and possibly weakness.[11][8][9][10]
Genetics
This disorder has been associated with several different mutations and is inherited in an autosomal recessive manner. It was originally believed to affect only those of northern Swedish ancestry, however the disease has been found in those of Norwegian and Finnish decent as well. The carrier rate in northern Sweden has been estimated at 1:188.[11] ISCU deficiency has been linked to pathogenic variants including intronic variants c.418+382G>C, g.7044G>C,[12] and IVS5+382 G>C[13] as well as a c.149G>A missense mutation in exon 3.[14] The intronic mutations have been suggested to activate a cryptic splice site, resulting in the production of a splice variant that encodes a putatively non-functional protein.[10]
Interactions
ISCU has been shown to have 235 binary protein-protein interactions including 79 co-complex interactions. ISCU appears to interact with ISCS, NUP62, SDHB, HPRT1, CCDC172, GOLGA2, IKZF1, KRT40, AGTRAP, NECAB2, FAM9B, BANP, LNX1, MID2, GOLGA6L9, ccdc136, KRT34, SPERT, PICK1, YWHAB, SFN, mbl, E7, dnaX, hscB, MAPk-Ak2, hale, and cv-c.[15]
References
- GRCh38: Ensembl release 89: ENSG00000136003 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000025825 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Hwang DM, Dempsey A, Tan KT, Liew CC (November 1996). "A modular domain of NifU, a nitrogen fixation cluster protein, is highly conserved in evolution". Journal of Molecular Evolution. 43 (5): 536–40. Bibcode:1996JMolE..43..536H. doi:10.1007/BF02337525. PMID 8875867. S2CID 21873104.
- Tong WH, Rouault T (November 2000). "Distinct iron-sulfur cluster assembly complexes exist in the cytosol and mitochondria of human cells". The EMBO Journal. 19 (21): 5692–700. doi:10.1093/emboj/19.21.5692. PMC 305809. PMID 11060020.
- "Entrez Gene: ISCU iron-sulfur cluster scaffold homolog (E. coli)".
 This article incorporates text from this source, which is in the public domain.
This article incorporates text from this source, which is in the public domain. - "ISCU - Iron-sulfur cluster assembly enzyme ISCU, mitochondrial precursor - Homo sapiens (Human) - ISCU gene & protein". www.uniprot.org. Retrieved 2018-09-04.
 This article incorporates text available under the CC BY 4.0 license.
This article incorporates text available under the CC BY 4.0 license. - "UniProt: the universal protein knowledgebase". Nucleic Acids Research. 45 (D1): D158–D169. January 2017. doi:10.1093/nar/gkw1099. PMC 5210571. PMID 27899622.
- Mochel F, Haller RG (2009-03-31). "Myopathy with Deficiency of ISCU – RETIRED CHAPTER, FOR HISTORICAL REFERENCE ONLY". Myopathy with Deficiency of ISCU. GeneReviews. PMID 20301757. Updated 2011 Sep 1
 This article incorporates text from this source, which is in the public domain.
This article incorporates text from this source, which is in the public domain. - Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K, Taivassalo T, Andersen PM, Singleton A, Rouault TA, Fischbeck KH, Haller RG (March 2008). "Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance". American Journal of Human Genetics. 82 (3): 652–60. doi:10.1016/j.ajhg.2007.12.012. PMC 2427212. PMID 18304497.
- Sanaker PS, Toompuu M, Hogan VE, He L, Tzoulis C, Chrzanowska-Lightowlers ZM, Taylor RW, Bindoff LA (June 2010). "Differences in RNA processing underlie the tissue specific phenotype of ISCU myopathy". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease. 1802 (6): 539–44. doi:10.1016/j.bbadis.2010.02.010. PMID 20206689.
- Kollberg G, Melberg A, Holme E, Oldfors A (February 2011). "Transient restoration of succinate dehydrogenase activity after rhabdomyolysis in iron-sulphur cluster deficiency myopathy". Neuromuscular Disorders. 21 (2): 115–20. doi:10.1016/j.nmd.2010.11.010. PMID 21196119. S2CID 38626230.
- Kollberg G, Tulinius M, Melberg A, Darin N, Andersen O, Holmgren D, Oldfors A, Holme E (August 2009). "Clinical manifestation and a new ISCU mutation in iron-sulphur cluster deficiency myopathy". Brain. 132 (Pt 8): 2170–9. doi:10.1093/brain/awp152. PMID 19567699.
- "235 binary interactions found for search term ISCU". IntAct Molecular Interaction Database. EMBL-EBI. Retrieved 2018-09-05.
Further reading
- Mochel F, Knight MA, Tong WH, Hernandez D, Ayyad K, Taivassalo T, Andersen PM, Singleton A, Rouault TA, Fischbeck KH, Haller RG (March 2008). "Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance". American Journal of Human Genetics. 82 (3): 652–60. doi:10.1016/j.ajhg.2007.12.012. PMC 2427212. PMID 18304497.
- Li K, Tong WH, Hughes RM, Rouault TA (May 2006). "Roles of the mammalian cytosolic cysteine desulfurase, ISCS, and scaffold protein, ISCU, in iron-sulfur cluster assembly". The Journal of Biological Chemistry. 281 (18): 12344–51. doi:10.1074/jbc.M600582200. PMID 16527810.
- Tong WH, Rouault TA (March 2006). "Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis". Cell Metabolism. 3 (3): 199–210. doi:10.1016/j.cmet.2006.02.003. PMID 16517407.
- Acquaviva F, De Biase I, Nezi L, Ruggiero G, Tatangelo F, Pisano C, Monticelli A, Garbi C, Acquaviva AM, Cocozza S (September 2005). "Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2". Journal of Cell Science. 118 (Pt 17): 3917–24. doi:10.1242/jcs.02516. PMID 16091420.
- Benzinger A, Muster N, Koch HB, Yates JR, Hermeking H (June 2005). "Targeted proteomic analysis of 14-3-3 sigma, a p53 effector commonly silenced in cancer". Molecular & Cellular Proteomics. 4 (6): 785–95. doi:10.1074/mcp.M500021-MCP200. PMID 15778465.
External links
- GeneReviews/NCBI/NIH/UW entry on Myopathy with Deficiency of ISCU
- GeneReviews/NIH/NCBI/UW entry on Myopathy with Deficiency of ISCU
This article incorporates text from the United States National Library of Medicine, which is in the public domain.