Transcription preinitiation complex
The preinitiation complex (abbreviated PIC) is a complex of approximately 100 proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The preinitiation complex positions RNA polymerase II at gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription.[1][2][3][4]
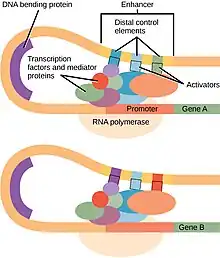
The minimal PIC includes RNA polymerase II and six general transcription factors: TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH. Additional regulatory complexes (such as the mediator coactivator[5] and chromatin remodeling complexes) may also be components of the PIC.
Assembly
A classical view of PIC formation at the promoter involves the following steps:
- TATA binding protein (TBP, a subunit of TFIID) binds the promoter, creating a sharp bend in the promoter DNA.
- TBP recruits TFIIA, then TFIIB, to the promoter.
- TFIIB recruits RNA polymerase II and TFIIF to the promoter.
- TFIIE joins the growing complex and recruits TFIIH which has protein kinase activity (phosphorylates RNA polymerase II within the CTD) and DNA helicase activity (unwinds DNA at promoter). It also recruits nucleotide-excision repair proteins.
- Subunits within TFIIH that have ATPase and helicase activity create negative superhelical tension in the DNA.
- Negative superhelical tension causes approximately one turn of DNA to unwind and form the transcription bubble.
- The template strand of the transcription bubble engages with the RNA polymerase II active site.
- RNA synthesis begins.
- After synthesis of ~10 nucleotides of RNA, and an obligatory phase of several abortive transcription cycles, RNA polymerase II escapes the promoter region to transcribe the remainder of the gene.
An alternative hypothesis of PIC assembly postulates the recruitment of a pre-assembled "RNA polymerase II holoenzyme" directly to the promoter (composed of all, or nearly all GTFs and RNA polymerase II and regulatory complexes), in a manner similar to the bacterial RNA polymerase (RNAP).
Other preinitiation complexes
Archaea have a preinitiation complex resembling that of a minimized Pol II PIC, with a TBP and an Archaeal transcription factor B (TFB, a TFIIB homolog). The assembly follows a similar sequence, starting with TBP binding to the promoter. An interesting aspect is that the entire complex is bound in an inverse orientation compared to those found in eukaryotic PIC.[7] They also use TFE, a TFIIE homolog, which assists in transcription initiation but is not required.[8][9]
Pol I initiation start with UBTF (UBF) recognizing an upstream control element (UCE) located around ~100 to 200 bp upstream. It recruits Selective factor 1 (TIF-IB), which is a complex of TBP and three units of TBP-associated factor. UBF then recognizes the core control elements. Phosphorylated RRN3 (TIF-IB) binds Pol I. The entire complex recognizes UBF/SL1, binds to it, and starts transcribing. The precise usage of subunits differ among organisms.[10]
Pol III has three classes of initiation, which start with different factors recognizing different control elements but all converging on TFIIIB (similar to TFIIB-TBP; consists of TBP/TRF, a TFIIB-related factor, and a B″ unit) recruiting the Pol III preinitiation complex. The overall architecture resembles that of Pol II. Only TFIIIB needs to remain attached during elongation.[11]
References
- Lee TI, Young RA (2000). "Transcription of eukaryotic protein-coding genes". Annual Review of Genetics. 34: 77–137. doi:10.1146/annurev.genet.34.1.77. PMID 11092823.
- Kornberg RD (August 2007). "The molecular basis of eukaryotic transcription". Proceedings of the National Academy of Sciences of the United States of America. 104 (32): 12955–61. Bibcode:2007PNAS..10412955K. doi:10.1073/pnas.0704138104. PMC 1941834. PMID 17670940.
- Kim TK, Lagrange T, Wang YH, Griffith JD, Reinberg D, Ebright RH (November 1997). "Trajectory of DNA in the RNA polymerase II transcription preinitiation complex". Proceedings of the National Academy of Sciences of the United States of America. 94 (23): 12268–73. Bibcode:1997PNAS...9412268K. doi:10.1073/pnas.94.23.12268. PMC 24903. PMID 9356438.
- Kim TK, Ebright RH, Reinberg D (May 2000). "Mechanism of ATP-dependent promoter melting by transcription factor IIH". Science. 288 (5470): 1418–22. Bibcode:2000Sci...288.1418K. doi:10.1126/science.288.5470.1418. PMID 10827951.
- Allen BL, Taatjes DJ (2015). "The Mediator complex: a central integrator of transcription". Nature Reviews. Molecular Cell Biology. 16 (3): 155–66. doi:10.1038/nrm3951. PMC 4963239. PMID 25693131.
- Duttke, SH (March 2015). "Evolution and diversification of the basal transcription machinery". Trends in Biochemical Sciences. 40 (3): 127–9. doi:10.1016/j.tibs.2015.01.005. PMC 4410091. PMID 25661246.
- Bell, SD; Jackson, SP (June 1998). "Transcription and translation in Archaea: a mosaic of eukaryal and bacterial features". Trends in Microbiology. 6 (6): 222–8. doi:10.1016/S0966-842X(98)01281-5. PMID 9675798.
- Hanzelka, BL; Darcy, TJ; Reeve, JN (March 2001). "TFE, an archaeal transcription factor in Methanobacterium thermoautotrophicum related to eucaryal transcription factor TFIIEalpha". Journal of Bacteriology. 183 (5): 1813–8. doi:10.1128/JB.183.5.1813-1818.2001. PMC 95073. PMID 11160119.
- Gehring, Alexandra M.; Walker, Julie E.; Santangelo, Thomas J.; Margolin, W. (15 July 2016). "Transcription Regulation in Archaea". Journal of Bacteriology. 198 (14): 1906–1917. doi:10.1128/JB.00255-16. PMC 4936096. PMID 27137495.
- Grummt, Ingrid (15 July 2003). "Life on a planet of its own: regulation of RNA polymerase I transcription in the nucleolus". Genes & Development. 17 (14): 1691–1702. doi:10.1101/gad.1098503R. PMID 12865296.
- Han, Y; Yan, C; Fishbain, S; Ivanov, I; He, Y (2018). "Structural visualization of RNA polymerase III transcription machineries". Cell Discovery. 4: 40. doi:10.1038/s41421-018-0044-z. PMC 6066478. PMID 30083386.
External links
- Descriptive image – biochem.ucl.ac.uk