Bunyamwera orthobunyavirus
Bunyamwera orthobunyavirus (BUNV) is a negative-sense, single-stranded enveloped RNA virus. It is assigned to the Orthobunyavirus genus, in the Bunyavirales order.
| Bunyamwera orthobunyavirus | |
|---|---|
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Class: | Ellioviricetes |
| Order: | Bunyavirales |
| Family: | Peribunyaviridae |
| Genus: | Orthobunyavirus |
| Species: | Bunyamwera orthobunyavirus |
| Isolates[1] | |
| |
| Synonyms[2] | |
| |
Bunyamwera orthobunyavirus can infect both humans and Aedes aegypti (yellow fever mosquito).[3]
It is named for Bunyamwera, a town in western Uganda, where it was isolated in 1943. Reassortant viruses derived from Bunyamwera orthobunyavirus, such as Ngari virus, have been associated with large outbreaks of viral haemorrhagic fever in Kenya and Somalia.[4][5][6]
Molecular biology
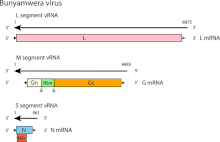
The genetic structure of Bunyamwera orthobunyavirus is typical for Bunyavirales viruses, which are an order of enveloped negative-sense, single-stranded RNA viruses with a genome split into three parts—Small (S), Middle (M), and Large (L). The L RNA segment encodes an RNA-dependent RNA polymerase (L protein), the M RNA segment encodes two surface glycoproteins (Gc and Gn) and a nonstructural protein (NSm), while the S RNA segment encodes a nucleocapsid protein (N) and, in an alternative overlapping reading frame, a second nonstructural protein (NSs).[7] The genomic RNA segments are encapsidated by copies of the N protein in the form of ribonucleoprotein (RNP) complexes.[8] The N protein is the most abundant protein in virus particles and infected cells and, therefore, the main target in many serological and molecular diagnostics.[9][10]
Disease in humans
| Bunyamwera fever | |
|---|---|
| Specialty | Infectious disease |
In humans, Bunyamwera orthobunyavirus causes Bunyamwera fever.
References
- "ICTV 9th Report (2011) Bunyaviridae" (html). International Committee on Taxonomy of Viruses (ICTV). Retrieved 31 January 2019.
Bunya: from Bunyamwera, place in Uganda, where type virus was isolated.
- "ICTV Taxonomy history: Bunyamwera orthobunyavirus" (html). International Committee on Taxonomy of Viruses (ICTV). Retrieved 31 January 2019.
- "Bunyamwera virus (BUNV)".
- Gerrard SR, Li L, Barrett AD, Nichol ST (2004). "Ngari virus is a Bunyamwera virus reassortant that can be associated with large outbreaks of hemorrhagic fever in Africa". J Virol. 78 (16): 8922–6. doi:10.1128/JVI.78.16.8922-8926.2004. PMC 479050. PMID 15280501.
- Odhiambo C, Venter M, Limbaso K, Swanepoel R, Sang R (2014). "Genome sequence analysis of in vitro and in vivo phenotypes of Bunyamwera and Ngari virus isolates from northern Kenya". PLOS ONE. 9 (8): e105446. Bibcode:2014PLoSO...9j5446O. doi:10.1371/journal.pone.0105446. PMC 4143288. PMID 25153316.
- Briese, T.; Bird, B.; Kapoor, V.; Nichol, S. T.; Lipkin, W. I. (12 May 2006). "Batai and Ngari Viruses: M Segment Reassortment and Association with Severe Febrile Disease Outbreaks in East Africa". Journal of Virology. 80 (11): 5627–5630. doi:10.1128/JVI.02448-05. PMC 1472162. PMID 16699043.
- Plyusnin, Alexander; Elliott, Richard M (2011-01-01). Bunyaviridae: molecular and cellular biology. Norfolk, UK: Caister Academic Press. ISBN 9781904455905. OCLC 711044654.
- Ariza, Antonio; Tanner, Sian J.; Walter, Cheryl T.; Dent, Kyle C.; Shepherd, Dale A.; Wu, Weining; Matthews, Susan V.; Hiscox, Julian A.; Green, Todd J. (2013-06-01). "Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization". Nucleic Acids Research. 41 (11): 5912–5926. doi:10.1093/nar/gkt268. ISSN 0305-1048. PMC 3675483. PMID 23595147.
- Bilk, S.; Schulze, C.; Fischer, M.; Beer, M.; Hlinak, A.; Hoffmann, B. (2012-09-14). "Organ distribution of Schmallenberg virus RNA in malformed newborns". Veterinary Microbiology. 159 (1–2): 236–238. doi:10.1016/j.vetmic.2012.03.035. PMID 22516190.
- Bréard, Emmanuel; Lara, Estelle; Comtet, Loïc; Viarouge, Cyril; Doceul, Virginie; Desprat, Alexandra; Vitour, Damien; Pozzi, Nathalie; Cay, Ann Brigitte (2013-01-15). "Validation of a Commercially Available Indirect Elisa Using a Nucleocapside Recombinant Protein for Detection of Schmallenberg Virus Antibodies". PLOS ONE. 8 (1): e53446. Bibcode:2013PLoSO...853446B. doi:10.1371/journal.pone.0053446. ISSN 1932-6203. PMC 3546048. PMID 23335964.
External links