Peptidoglycan
Peptidoglycan or murein is a polysaccharide consisting of sugars and amino acids that forms a mesh-like peptidoglycan layer outside the plasma membrane, the rigid cell wall (murein sacculus) characteristic of most bacteria (domain Bacteria[1]). The sugar component consists of alternating residues of β-(1,4) linked N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM). Attached to the N-acetylmuramic acid is a oligopeptide chain made of three to five amino acids. The peptide chain can be cross-linked to the peptide chain of another strand forming the 3D mesh-like layer.[2] Peptidoglycan serves a structural role in the bacterial cell wall, giving structural strength, as well as counteracting the osmotic pressure of the cytoplasm. This repetitive linking results in a dense peptidoglycan layer. This peptidoglycan layer is critical for maintaining cell form and withstanding high osmotic pressures, and it is regularly replaced by peptidoglycan production. Peptidoglycan hydrolysis and synthesis are two processes that must occur in order for cells to grow and multiply. This technique is carried out in three stages: clipping of current material, insertion of new material, and re-crosslinking of existing material to new material.[3]
The peptidoglycan layer is substantially thicker in Gram-positive bacteria (20 to 80 nanometers) than in Gram-negative bacteria (7 to 8 nanometers).[4] Depending on pH growth conditions, the peptidoglycan forms around 40 to 90% of the cell wall's dry weight of Gram-positive bacteria but only around 10% of Gram-negative strains. Thus, presence of high levels of peptidoglycan is the primary determinant of the characterisation of bacteria as Gram-positive.[5] In Gram-positive strains, it is important in attachment roles and serotyping purposes.[6] For both Gram-positive and Gram-negative bacteria, particles of approximately 2 nm can pass through the peptidoglycan.[7]
It is difficult to tell whether an organism is gram-positive or gram-negative using a microscope; Gram staining, created by Hans Christian Gram in 1884, is required. The bacteria are stained with several dyes such as crystal violet, iodine alcohol, and safranin using the gram staining procedure. Gram positive cells are purple after staining, while Gram negative cells are red.[8]
Structure and importance
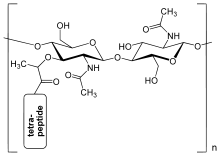
The peptidoglycan layer in the bacterial cell wall is a crystal lattice structure formed from linear chains of two alternating amino sugars, namely N-acetylglucosamine (GlcNAc or NAGA) and N-acetylmuramic acid (MurNAc or NAMA). The alternating sugars are connected by a β-(1,4)-glycosidic bond. Each MurNAc is attached to a short (4- to 5-residue) amino acid chain, containing L-alanine, D-glutamic acid, meso-diaminopimelic acid, and D-alanine in the case of Escherichia coli (a Gram-negative bacterium) or L-alanine, D-glutamine, L-lysine, and D-alanine with a 5-glycine interbridge between tetrapeptides in the case of Staphylococcus aureus (a Gram-positive bacterium). Peptidoglycan is one of the most important sources of D-amino acids in nature.
By enclosing the inner membrane, the peptididoglycan layer protects the cell from lysis caused by the turgor pressure of the cell. When the cell wall grows, it retains its shape throughout its life, so a rod shape will remain a rod shape, and a spherical shape will remain a spherical shape for life. This happens because the freshly added septal material of synthesis transforms into a hemispherical wall for the offspring cells.[9]
Cross-linking between amino acids in different linear amino sugar chains occurs with the help of the enzyme DD-transpeptidase and results in a 3-dimensional structure that is strong and rigid. The specific amino acid sequence and molecular structure vary with the bacterial species. [10]
The different peptidoglycan types of bacterial cell walls and their taxonomic implications have been described.[11] Archaea (domain Archaea[1]) do not contain peptidoglycan (murein).[12] Some Archaea contain pseudopeptidoglycan (pseudomurein, see below).[13]
 The structure of peptidoglycan. NAG = N-acetylglucosamine (also called GlcNAc or NAGA), NAM = N-acetylmuramic acid (also called MurNAc or NAMA).
The structure of peptidoglycan. NAG = N-acetylglucosamine (also called GlcNAc or NAGA), NAM = N-acetylmuramic acid (also called MurNAc or NAMA).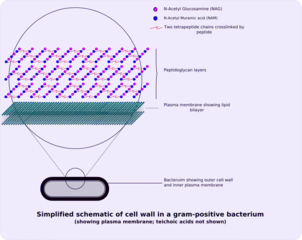 Gram-positive cell wall
Gram-positive cell wall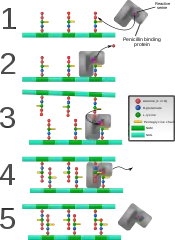 Penicillin binding protein forming cross-links in newly formed bacterial cell wall.
Penicillin binding protein forming cross-links in newly formed bacterial cell wall.
Peptidoglycan is involved in binary fission during bacterial cell reproduction. L-form bacteria and mycoplasmas, both lacking peptidoglycan cell walls, do not proliferate by binary fission, but by a budding mechanism.[14][15]
In the course of early evolution, the successive development of boundaries (membranes, walls) protecting first structures of life against their environment must have been essential for the formation of the first cells (cellularisation).
The invention of rigid peptidoglycan (murein) cell walls in bacteria (domain Bacteria[1]) was probably the prerequisite for their survival, extensive radiation and colonisation of virtually all habitats of the geosphere and hydrosphere.[16][17]
Biosynthesis
The peptidoglycan monomers are synthesized in the cytosol and are then attached to a membrane carrier bactoprenol. Bactoprenol transports peptidoglycan monomers across the cell membrane where they are inserted into the existing peptidoglycan.[18]
- In the first step of peptidoglycan synthesis, glutamine, which is an amino acid, donates an amino group to a sugar, fructose 6-phosphate.[19] This reaction, catalyzed by EC 2.6.1.16 (GlmS), turns fructose 6-phosphate into glucosamine-6-phosphate.[20]
- In step two, an acetyl group is transferred from acetyl CoA to the amino group on the glucosamine-6-phosphate creating N-acetyl-glucosamine-6-phosphate.[19] This reaction is EC 5.4.2.10, catalyzed by GlmM.[20]
- In step three of the synthesis process, the N-acetyl-glucosamine-6-phosphate is isomerized, which will change N-acetyl-glucosamine-6-phosphate to N-acetyl-glucosamine-1-phosphate.[19] This is EC 2.3.1.157, catalyzed by GlmU.[20]
- In step 4, the N-acetyl-glucosamine-1-phosphate, which is now a monophosphate, attacks UTP. Uridine triphosphate, which is a pyrimidine nucleotide, has the ability to act as an energy source. In this particular reaction, after the monophosphate has attacked the UTP, an inorganic pyrophosphate is given off and is replaced by the monophosphate, creating UDP-N-acetylglucosamine (2,4). (When UDP is used as an energy source, it gives off an inorganic phosphate.) This initial stage, is used to create the precursor for the NAG in peptidoglycan.[19] This is EC 2.7.7.23, also catalyzed by GlmU, which is a bifunctional enzyme.[20]
- In step 5, some of the UDP-N-acetylglucosamine (UDP-GlcNAc) is converted to UDP-MurNAc (UDP-N-acetylmuramic acid) by the addition of a lactyl group to the glucosamine. Also in this reaction, the C3 hydroxyl group will remove a phosphate from the alpha carbon of phosphoenolpyruvate. This creates what is called an enol derivative.[19] EC 2.5.1.7, catalyzed by MurA.[20]
- In step 6, the enol is reduced to a “lactyl moiety” by NADPH in step six.[19] EC 1.3.1.98, catalyzed by MurB.[20]
- In step 7, the UDP–MurNAc is converted to UDP-MurNAc pentapeptide by the addition of five amino acids, usually including the dipeptide D-alanyl-D-alanine.[19] This is a string of three reactions: EC 6.3.2.8 by MurC, EC 6.3.2.9 by MurD, and EC 6.3.2.13 by MurE.[20]
Each of these reactions requires the energy source ATP.[19] This is all referred to as Stage one.
Stage two occurs in the cytoplasmic membrane. It is in the membrane where a lipid carrier called bactoprenol carries peptidoglycan precursors through the cell membrane.
- Undecaprenyl phosphate will attack the UDP-MurNAc penta, creating a PP-MurNac penta, which is now a lipid (lipid I).[19] EC 2.7.8.13 by MraY.[20]
- UDP-GlcNAc is then transported to MurNAc, creating Lipid-PP-MurNAc penta-GlcNAc (lipid II), a disaccharide, also a precursor to peptidoglycan.[19] EC 2.4.1.227 by MurG.[20]
- Lipid II is transported across the membrane by flippase (MurJ), a discovery made in 2014 after decades of searching.[21] Once it is there, it is added to the growing glycan chain by the enzyme peptidoglycan glycosyltransferase (GTase, EC 2.4.1.129). This reaction is known as tranglycosylation. In the reaction, the hydroxyl group of the GlcNAc will attach to the MurNAc in the glycan, which will displace the lipid-PP from the glycan chain.[19]
- In a final step, the DD-transpeptidase (TPase, EC 3.4.16.4) crosslins individual glycan chains. This protein is also known as the penicillin-binding protein. Some versions of the enzyme also performs the glycosyltransferase function, while others leave the job to a separate enzyme.[20]
Inhibition
Some antibacterial drugs such as penicillin interfere with the production of peptidoglycan by binding to bacterial enzymes known as penicillin-binding proteins or DD-transpeptidases.[6] Penicillin-binding proteins form the bonds between oligopeptide crosslinks in peptidoglycan. For a bacterial cell to reproduce through binary fission, more than a million peptidoglycan subunits (NAM-NAG+oligopeptide) must be attached to existing subunits.[22] Mutations in genes coding for transpeptidases that lead to reduced interactions with an antibiotic are a significant source of emerging antibiotic resistance.[23] Since peptidoglycan is also lacking in L-form bacteria and in mycoplasmas, both are resistant against penicillin.
Other steps of peptidoglycan synthesis can also be targeted. The topical antibiotic bacitracin targets the utilization of C55-isoprenyl pyrophosphate. Lantibiotics, which includes the food preservative nisin, attack lipid II.[24]
Lysozyme, which is found in tears and constitutes part of the body's innate immune system exerts its antibacterial effect by breaking the β-(1,4)-glycosidic bonds in peptidoglycan (see above).
Pseudopeptidoglycan (Pseudomurein)
In some archaea, i.e. members of the Methanobacteriales and in the genus Methanopyrus, pseudopeptidoglycan (pseudomurein) has been found.[13] In pseudopeptidoglycan the sugar residues are β-(1,3) linked N-acetylglucosamine and N-acetyltalosaminuronic acid. This makes the cell walls of such archaea insensitive to lysozyme.[25] The biosynthesis of pseudopeptidoglycan has been described.[26]
See also
- Undecaprenyl-diphosphatase
References
- Woese, Carl R.; Wheelis, Mark L.; Kandler, Otto (June 1990). "Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya" (PDF). Proc Natl Acad Sci USA. 87 (12): 4576–4579. Bibcode:1990PNAS...87.4576W. doi:10.1073/pnas.87.12.4576. PMC 54159. PMID 2112744.
- Mehta A (20 March 2011). "Animation of Synthesis of Peptidoglycan Layer". PharmaXChange.info.
- Belgrave AM, Wolgemuth CW (June 2013). "Elasticity and biochemistry of growth relate replication rate to cell length and cross-link density in rod-shaped bacteria". Biophysical Journal. 104 (12): 2607–2611. Bibcode:2013BpJ...104.2607B. doi:10.1016/j.bpj.2013.04.028. PMC 3686348. PMID 23790368.
- Purcell A (18 March 2016). "Bacteria". Basic Biology.
- Hogan CM (12 October 2014). "Bacteria". In Draggan S, Cleveland CJ (eds.). Encyclopedia of Earth. Washington DC: National Council for Science and the Environment.
- Salton MR, Kim KS (1996). "Structure". In Baron S, et al. (eds.). Structure. In: Baron's Medical Microbiology (4th ed.). Univ of Texas Medical Branch. ISBN 978-0-9631172-1-2. PMID 21413343.
- Demchick P, Koch AL (February 1996). "The permeability of the wall fabric of Escherichia coli and Bacillus subtilis". Journal of Bacteriology. 178 (3): 768–773. doi:10.1128/jb.178.3.768-773.1996. PMC 177723. PMID 8550511.
- Coico R (October 2005). "Gram Staining". In Coico R, Kowalik T, Quarles J, Stevenson B (eds.). Current Protocols in Microbiology. Vol. Appendix 3. Hoboken, NJ, USA: John Wiley & Sons, Inc. pp. mca03cs00. doi:10.1002/9780471729259.mca03cs00. ISBN 978-0-471-72925-9. PMID 18770544. S2CID 32452815.
- Huang KC, Mukhopadhyay R, Wen B, Gitai Z, Wingreen NS (December 2008). "Cell shape and cell-wall organization in Gram-negative bacteria". Proceedings of the National Academy of Sciences of the United States of America. 105 (49): 19282–19287. Bibcode:2008PNAS..10519282H. doi:10.1073/pnas.0805309105. PMC 2592989. PMID 19050072.
- Ryan KJ, Ray CG, eds. (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. ISBN 978-0-8385-8529-0.
- Schleifer KH, Kandler O (December 1972). "Peptidoglycan types of bacterial cell walls and their taxonomic implications". Bacteriological Reviews. 36 (4): 407–477. doi:10.1128/MMBR.36.4.407-477.1972. PMC 408328. PMID 4568761.
- Kandler O, Hippe H (May 1977). "Lack of peptidoglycan in the cell walls of Methanosarcina barkeri". Archives of Microbiology. 113 (1–2): 57–60. doi:10.1007/BF00428580. PMID 889387. S2CID 19145374.
- Kandler O, König H (April 1998). "Cell wall polymers in Archaea (Archaebacteria)". Cellular and Molecular Life Sciences. 54 (4): 305–308. doi:10.1007/s000180050156. PMID 9614965. S2CID 13527169.
- Kandler, Gertraud; Kandler, Otto (1954). (Article in English available). "Untersuchungen über die Morphologie und die Vermehrung der pleuropneumonie-ähnlichen Organismen und der L-Phase der Bakterien. I. Lichtmikroskopische Untersuchungen" [Studies on morphology and multiplication of pleuropneumonia-like organisms and on bacterial L-phase, I. Light microscopy (now mycoplasmas and L-form bacteria)] (PDF). Archiv für Mikrobiologie (in German). 21 (2): 178–201. doi:10.1007/BF01816378. PMID 14350641. S2CID 21257985.
- Leaver, M.; Domínguez-Cuevas, P.; Coxhead, J. M.; Daniel, R. A.; Errington, Jeff (1 February 2009). [see also Erratum, 23 July 2009, Nature, vol. 460, p.538]. "Life without a wall or division machine in Bacillus subtilis". Nature. 457: 849–853. doi:10.1038/nature07742.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Kandler, Otto (1994). "The early diversification of life". In Bengtson, Stefan (ed.). Early Life on Earth. Nobel Symposium 84. New York: Columbia U.P. pp. 221–270. ISBN 9780231080880.
- Kandler, Otto (1998). "The early diversification of life and the origin of the three domains: A proposal". In Wiegel, Jürgen; Adams, Michael W.W. (eds.). Thermophiles: The keys to molecular evolution and the origin of life?. London: Taylor and Francis Ltd. pp. 19–31. ISBN 978-0-203-48420-3.
- "The Prokaryotic Cell: Bacteria". Archived from the original on 26 July 2010. Retrieved 1 May 2011.
- White D (2007). The physiology and biochemistry of prokaryotes (3rd ed.). NY: Oxford University Press Inc.
- Otten, Christian; Brilli, Matteo; Vollmer, Waldemar; Viollier, Patrick H.; Salje, Jeanne (January 2018). "Peptidoglycan in obligate intracellular bacteria: Peptidoglycan in obligate intracellular bacteria". Molecular Microbiology. 107 (2): 142–163. doi:10.1111/mmi.13880.
- Lok-To Sham; Emily K. Butler; Matthew D. Lebar; et al. (11 July 2014). "MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis". Science. 345 (6193): 220–222. Bibcode:2014Sci...345..220S. doi:10.1126/science.1254522. PMC 4163187. PMID 25013077.
- Bauman R (2007). 2nd (ed.). Microbiology with Diseases by Taxonomy. Benjamin Cummings. ISBN 978-0-8053-7679-1.
- Spratt BG (April 1994). "Resistance to antibiotics mediated by target alterations". Science. 264 (5157): 388–393. Bibcode:1994Sci...264..388S. doi:10.1126/science.8153626. PMID 8153626. S2CID 30578841.
- Sarkar, P; Yarlagadda, V; Ghosh, C; Haldar, J (1 March 2017). "A review on cell wall synthesis inhibitors with an emphasis on glycopeptide antibiotics". MedChemComm. 8 (3): 516–533. doi:10.1039/c6md00585c. PMC 6072328. PMID 30108769.
- Madigan MT, Martinko JM, Dunlap PV, Clark DP (2009). Brock Biology of Microorganisms (12th ed.). San Francisco, CA: Pearson/Benjamin Cummings.
- König H, Kandler O, Hammes W (January 1989). "Biosynthesis of pseudomurein: isolation of putative precursors from Methanobacterium thermoautotrophicum". Canadian Journal of Microbiology. 35 (1): 176–181. doi:10.1139/m89-027. PMID 2720492.