Bacillota
The Bacillota (synonym Firmicutes) are a phylum of bacteria, most of which have gram-positive cell wall structure.[2] The renaming of phyla such as Firmicutes in 2021 remains controversial among microbiologists, many of whom continue to use the earlier names of long standing in the literature.[3]
| Bacillota | |
|---|---|
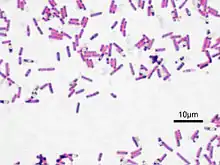 | |
| Bacillus subtilis, Gram-stained | |
| Scientific classification | |
| Domain: | Bacteria |
| (unranked): | Terrabacteria |
| Phylum: | Bacillota Gibbons and Murray 2021[1] |
| Classes | |
| |
| Synonyms | |
| |
The name "Firmicutes" was derived from the Latin words for "tough skin," referring to the thick cell wall typical of bacteria in this phylum. Scientists once classified the Firmicutes to include all gram-positive bacteria, but have recently defined them to be of a core group of related forms called the low-G+C group, in contrast to the Actinomycetota. They have round cells, called cocci (singular coccus), or rod-like forms (bacillus). A few Firmicutes, such as Megasphaera, Pectinatus, Selenomonas and Zymophilus, have a porous pseudo-outer membrane that causes them to stain gram-negative.
Many Bacillota (Firmicutes) produce endospores, which are resistant to desiccation and can survive extreme conditions. They are found in various environments, and the group includes some notable pathogens. Those in one family, the heliobacteria, produce energy through anoxygenic photosynthesis. Bacillota play an important role in beer, wine, and cider spoilage.
Classes
The group is typically divided into the Clostridia, which are anaerobic, and the Bacilli, which are obligate or facultative aerobes.
On phylogenetic trees, the first two groups show up as paraphyletic or polyphyletic, as do their main genera, Clostridium and Bacillus.[4] However, Bacillota as a whole is generally believed to be monophyletic, or paraphyletic with the exclusion of Mollicutes.[5]
Phylogeny
The currently accepted taxonomy based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[6] and the National Center for Biotechnology Information (NCBI).[7]
| 16S rRNA based LTP_01_2022[8][9][10] | GTDB 07-RS207 by Genome Taxonomy Database[11][12][13] | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
♦ Paraphyletic Firmicutes |
|
Genera
More than 274 genera were considered as of 2016 to be within the Bacillota phylum, notable genera of Bacillota include:
Bacilli, order Bacillales
Bacilli, order Lactobacillales
Clinical significance
Bacillota make up ~30% of the mouse and human gut microbiome.[14] The phylum Bacillota as part of the gut microbiota has been shown to be involved in energy resorption, and potentially related to the development of diabetes and obesity.[15][16][17][18] Within the gut of healthy human adults, the most abundant bacterium: Faecalibacterium prausnitzii (F. prausnitzii), which makes up 5% of the total gut microbiome, is a member of the Bacillota phylum. This species is directly associated with reduced low-grade inflammation in obesity.[19] F. prausnitzii has been found in higher levels within the guts of obese children than in non-obese children.
In multiple studies a higher abundance of Bacillota has been found in obese individuals than in lean controls. A higher level of Lactobacillus (of the Bacillota phylum) has been found in obese patients and in one study, obese patients put on weight loss diets showed a reduced amount of Bacillota within their guts.[20]
Diet changes in mice have also been shown to promote changes in Bacillota abundance. A higher relative abundance of Bacillota was seen in mice fed a western diet (high fat/high sugar) than in mice fed a standard low fat/ high polysaccharide diet. The higher amount of Bacillota was also linked to more adiposity and body weight within mice.[21] Specifically, within obese mice, the class Mollicutes (within the Bacillota phylum) was the most common. When the microbiota of obese mice with this higher Bacillota abundance was transplanted into the guts of germ-free mice, the germ-free mice gained a significant amount of fat as compared to those transplanted with the microbiota of lean mice with lower Bacillota abundance.[22]
The presence of Christensenella (Bacillota, in class Clostridia), isolated from human faeces, has been found to correlate with lower body mass index.[23]
References
- Oren A, Garrity GM (2021). "Valid publication of the names of forty-two phyla of prokaryotes". Int J Syst Evol Microbiol. 71 (10): 5056. doi:10.1099/ijsem.0.005056. PMID 34694987. S2CID 239887308.
- "Firmicutes" at Dorland's Medical Dictionary
- Robitzki, Dan (4 January 2022). "Newly Renamed Prokaryote Phyla Cause Uproar". The Scientist Magazine. Retrieved 23 May 2022.
- Wolf M, Müller T, Dandekar T, Pollack JD (May 2004). "Phylogeny of Firmicutes with special reference to Mycoplasma (Mollicutes) as inferred from phosphoglycerate kinase amino acid sequence data". Int. J. Syst. Evol. Microbiol. (Comparative Study). 54 (Pt 3): 871–5. CiteSeerX 10.1.1.126.3863. doi:10.1099/ijs.0.02868-0. PMID 15143038. Archived from the original on 2012-12-09.
- Ciccarelli, FD (2006). "Toward automatic reconstruction of a highly resolved tree of life". Science. 311 (5765): 1283–1287. Bibcode:2006Sci...311.1283C. CiteSeerX 10.1.1.381.9514. doi:10.1126/science.1123061. PMID 16513982. S2CID 1615592.
- J. P. Euzéby. "Firmicutes". List of Prokaryotic names with Standing in Nomenclature (LPSN). Archived from the original on January 27, 2013. Retrieved 2013-03-20.
- Sayers; et al. "Firmicutes". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 24 April 2019.
- "The LTP". Retrieved 20 June 2022.
- "LTP_all tree in newick format". Retrieved 20 June 2022.
- "LTP_01_2022 Release Notes" (PDF). Retrieved 20 June 2022.
- "GTDB release 07-RS207". Genome Taxonomy Database. Retrieved 20 June 2022.
- "bac120_r207.sp_labels". Genome Taxonomy Database. Retrieved 20 June 2022.
- "Taxon History". Genome Taxonomy Database. Retrieved 20 June 2022.
- Ley RE, Peterson DA, Gordon JI (2006). "Ecological and evolutionary forces shaping microbial diversity in the human intestine". Cell (Review). 124 (4): 837–848. doi:10.1016/j.cell.2006.02.017. PMID 16497592. S2CID 17203181.
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI (2006). "Microbial ecology: human gut microbes associated with obesity". Nature (Clinical Trial). 444 (7122): 1022–1023. Bibcode:2006Natur.444.1022L. doi:10.1038/4441022a. PMID 17183309. S2CID 205034045.
- Henig, Robin Marantz (2006-08-13). "Fat Factors". New York Times Magazine. Retrieved 2008-09-28.
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI (August 2005). "Obesity alters gut microbial ecology". Proc. Natl. Acad. Sci. USA (Research Support). 102 (31): 11070–11075. Bibcode:2005PNAS..10211070L. doi:10.1073/pnas.0504978102. PMC 1176910. PMID 16033867.
- Komaroff AL. The Microbiome and Risk for Obesity and Diabetes. JAMA. Published online December 22, 2016. doi:10.1001/jama.2016.20099
- Chakraborti, Chandra Kanti (15 November 2015). "New-found link between microbiota and obesity". World Journal of Gastrointestinal Pathophysiology. 6 (4): 110–119. doi:10.4291/wjgp.v6.i4.110. PMC 4644874. PMID 26600968.
- Million, M.; Lagier, J.-C; Yahav, D.; Paul, M. (April 2013). "Gut bacterial microbiota and obesity". Clinical Microbiology and Infection. 19 (4): 305–313. doi:10.1111/1469-0691.12172. PMID 23452229.
- Turnbaugh, Peter J. (17 April 2008). "Diet-Induced Obesity Is Linked to Marked but Reversible Alterations in the Mouse Distal Gut Microbiome". Cell Host & Microbe. 3 (4): 213–223. doi:10.1016/j.chom.2008.02.015. PMC 3687783. PMID 18407065.
- Million, M. (April 2013). "Gut bacterial microbiota and obesity". Cell Microbiology and Infection. 19 (4): 305–313. doi:10.1111/1469-0691.12172. PMID 23452229.
- Goodrich, Julia K.; Waters, Jillian L.; Poole, Angela C.; Sutter, Jessica L.; Koren, Omry; Blekhman, Ran; Beaumont, Michelle; Van Treuren, William; Knight, Rob; Bell, Jordana T.; Spector, Timothy D.; Clark, Andrew G.; Ley, Ruth E. (2014). "Human Genetics Shape the Gut Microbiome". Cell. 159 (4): 789–799. doi:10.1016/j.cell.2014.09.053. ISSN 0092-8674. PMC 4255478. PMID 25417156.

External links
- Phylum "Firmicutes" - J.P. Euzéby: List of Prokaryotic names with Standing in Nomenclature