CYP2A6
Cytochrome P450 2A6 (abbreviated CYP2A6) is a member of the cytochrome P450 mixed-function oxidase system, which is involved in the metabolism of xenobiotics in the body. CYP2A6 is the primary enzyme responsible for the oxidation of nicotine and cotinine. It is also involved in the metabolism of several pharmaceuticals, carcinogens, and a number of coumarin-type alkaloids. CYP2A6 is the only enzyme in the human body that appreciably catalyzes the 7-hydroxylation of coumarin, such that the formation of the product of this reaction, 7-hydroxycoumarin, is used as a probe for CYP2A6 activity.
The CYP2A6 gene is part of a large cluster of cytochrome P450 genes from the CYP2A, CYP2B and CYP2F subfamilies on chromosome 19q. The gene was formerly referred to as CYP2A3; however, it has been renamed CYP2A6.[4]
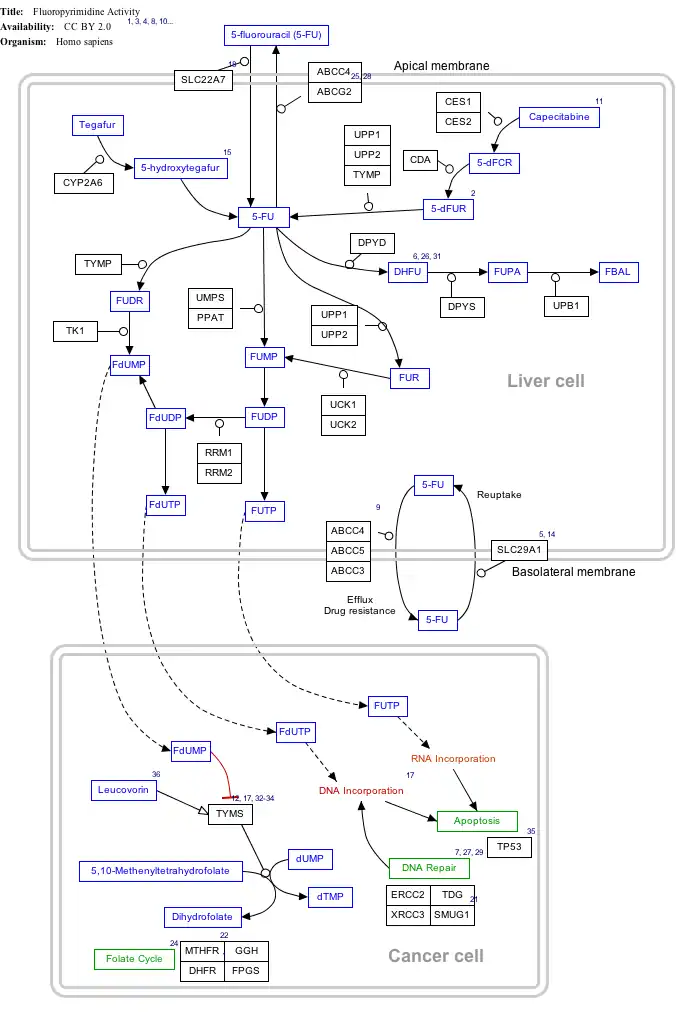
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles.[§ 1]
- ↑ The interactive pathway map can be edited at WikiPathways: "FluoropyrimidineActivity_WP1601".
Distribution
CYP2A6 localizes to the endoplasmic reticulum and is found predominantly in the liver.
Variability
Significant interindividual variability in CYP2A6 apoprotein and mRNA levels has been observed.
Induction
CYP2A6 is known to be inducible by phenobarbital and rifampicin, and it is suspected that other antiepileptic drugs may also have this effect.
CYP2A6 Ligands
| Substrates | Inhibitors | Inducers |
|---|---|---|
|
|
|
See also
References
- 1 2 3 GRCh38: Ensembl release 89: ENSG00000255974 - Ensembl, May 2017
- ↑ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ↑ "Entrez Gene: CYP2A6 cytochrome P450, family 2, subfamily A, polypeptide 6".
- 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 Lacy CF, Armstrong LL, Goldman MP, Lance LL (2007). Cytochrome P450 Enzymes: Substrates, Inhibitors, and Inducers. Hudson, OH: LexiComp Inc. pp. 1899–1912.
- ↑ Wen X, Wang JS, Neuvonen PJ, Backman JT (2002). "Isoniazid is a mechanism-based inhibitor of cytochrome P450 1A2, 2A6, 2C19 and 3A4 isoforms in human liver microsomes". Eur. J. Clin. Pharmacol. 57 (11): 799–804. doi:10.1007/s00228-001-0396-3. PMID 11868802. S2CID 19299097.
- ↑ Siu EC, Tyndale RF (2008). "Selegiline is a mechanism-based inactivator of CYP2A6 inhibiting nicotine metabolism in humans and mice". J. Pharmacol. Exp. Ther. 324 (3): 992–9. doi:10.1124/jpet.107.133900. PMID 18065502. S2CID 9833233.
Further reading
- Pelkonen O, Rautio A, Raunio H, Pasanen M (2000). "CYP2A6: a human coumarin 7-hydroxylase". Toxicology. 144 (1–3): 139–47. doi:10.1016/S0300-483X(99)00200-0. PMID 10781881.
- Fernandez-Salguero P, Gonzalez FJ (1995). "The CYP2A gene subfamily: species differences, regulation, catalytic activities and role in chemical carcinogenesis". Pharmacogenetics. 5 (Special Issue): S123–8. doi:10.1097/00008571-199512001-00013. PMID 7581481.
- Smith G, Stubbins MJ, Harries LW, Wolf CR (1999). "Molecular genetics of the human cytochrome P450 monooxygenase superfamily". Xenobiotica. 28 (12): 1129–65. doi:10.1080/004982598238868. PMID 9890157.
- Pelkonen O, Rautio A, Raunio H, Pasanen M (2000). "CYP2A6: a human coumarin 7-hydroxylase". Toxicology. 144 (1–3): 139–47. doi:10.1016/S0300-483X(99)00200-0. PMID 10781881.
- Tyndale RF, Sellers EM (2002). "Genetic variation in CYP2A6-mediated nicotine metabolism alters smoking behavior". Therapeutic Drug Monitoring. 24 (1): 163–71. doi:10.1097/00007691-200202000-00026. PMID 11805739. S2CID 12837210.
- Xu C, Goodz S, Sellers EM, Tyndale RF (2003). "CYP2A6 genetic variation and potential consequences". Adv. Drug Deliv. Rev. 54 (10): 1245–56. doi:10.1016/S0169-409X(02)00065-0. PMID 12406643.
- Kamataki T, Fujieda M, Kiyotani K, et al. (2005). "Genetic polymorphism of CYP2A6 as one of the potential determinants of tobacco-related cancer risk". Biochem. Biophys. Res. Commun. 338 (1): 306–10. doi:10.1016/j.bbrc.2005.08.268. PMID 16176798.
- Maurice M, Emiliani S, Dalet-Beluche I, et al. (1991). "Isolation and characterization of a cytochrome P450 of the IIA subfamily from human liver microsomes". Eur. J. Biochem. 200 (2): 511–7. doi:10.1111/j.1432-1033.1991.tb16212.x. PMID 1889415.
- Yun CH, Shimada T, Guengerich FP (1991). "Purification and characterization of human liver microsomal cytochrome P-450 2A6". Mol. Pharmacol. 40 (5): 679–85. PMID 1944238.
- Yamano S, Tatsuno J, Gonzalez FJ (1990). "The CYP2A3 gene product catalyzes coumarin 7-hydroxylation in human liver microsomes". Biochemistry. 29 (5): 1322–9. doi:10.1021/bi00457a031. PMID 2322567.
- Miles JS, Bickmore W, Brook JD, et al. (1989). "Close linkage of the human cytochrome P450IIA and P450IIB gene subfamilies: implications for the assignment of substrate specificity". Nucleic Acids Res. 17 (8): 2907–17. doi:10.1093/nar/17.8.2907. PMC 317701. PMID 2726448.
- Yamano S, Nagata K, Yamazoe Y, et al. (1989). "cDNA and deduced amino acid sequences of human P450 IIA3 (CYP2A3)". Nucleic Acids Res. 17 (12): 4888. doi:10.1093/nar/17.12.4888. PMC 318052. PMID 2748347.
- Phillips IR, Shephard EA, Ashworth A, Rabin BR (1985). "Isolation and sequence of a human cytochrome P-450 cDNA clone". Proc. Natl. Acad. Sci. U.S.A. 82 (4): 983–7. Bibcode:1985PNAS...82..983P. doi:10.1073/pnas.82.4.983. PMC 397177. PMID 3856261.
- Wainwright BJ, Watson EK, Shephard EA, Phillips IR (1985). "RFLP for a human cytochrome P-450 gene at 19q13.1-qter (HGM8 provisional designation CYPI)". Nucleic Acids Res. 13 (12): 4610. doi:10.1093/nar/13.12.4610. PMC 321810. PMID 4011450.
- Fernandez-Salguero P, Hoffman SM, Cholerton S, et al. (1995). "A genetic polymorphism in coumarin 7-hydroxylation: sequence of the human CYP2A genes and identification of variant CYP2A6 alleles". Am. J. Hum. Genet. 57 (3): 651–60. PMC 1801261. PMID 7668294.
- Ding S, Lake BG, Friedberg T, Wolf CR (1995). "Expression and alternative splicing of the cytochrome P-450 CYP2A7". Biochem. J. 306. ( Pt 1): 161–6. doi:10.1042/bj3060161. PMC 1136496. PMID 7864805.
- Hoffman SM, Fernandez-Salguero P, Gonzalez FJ, Mohrenweiser HW (1996). "Organization and evolution of the cytochrome P450 CYP2A-2B-2F subfamily gene cluster on human chromosome 19". J. Mol. Evol. 41 (6): 894–900. doi:10.1007/bf00173169. PMID 8587134. S2CID 12153961.
- Hadidi H, Zahlsen K, Idle JR, Cholerton S (1998). "A single amino acid substitution (Leu160His) in cytochrome P450 CYP2A6 causes switching from 7-hydroxylation to 3-hydroxylation of coumarin". Food Chem. Toxicol. 35 (9): 903–7. doi:10.1016/S0278-6915(97)00066-5. PMID 9409631.
- Oscarson M, Gullstén H, Rautio A, et al. (1998). "Genotyping of human cytochrome P450 2A6 (CYP2A6), a nicotine C-oxidase". FEBS Lett. 438 (3): 201–5. doi:10.1016/S0014-5793(98)01297-6. PMID 9827545. S2CID 1250823.
External links
- Human CPA6 genome location and CPA6 gene details page in the UCSC Genome Browser.
- Human CYP2A6 genome location and CYP2A6 gene details page in the UCSC Genome Browser.