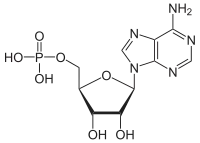
Adenosine monophosphate
 | |
 | |
| Names | |
|---|---|
| IUPAC name
5′-Adenylic acid | |
| Systematic IUPAC name
[(2R,3S,4R,5R)-5-(6-Amino-9H-purin-9-yl)-3,4-dihydroxyoxolan-2-yl]methyl dihydrogen phosphate | |
| Other names
Adenosine 5'-monophosphate | |
| Identifiers | |
3D model (JSmol) |
|
| ChEBI | |
| ChEMBL | |
| ChemSpider | |
| DrugBank | |
| ECHA InfoCard | 100.000.455 |
| KEGG | |
| MeSH | Adenosine+monophosphate |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C10H14N5O7P | |
| Molar mass | 347.22 g/mol |
| Appearance | white crystalline powder |
| Density | 2.32 g/mL |
| Melting point | 178 to 185 °C (352 to 365 °F; 451 to 458 K) |
| Boiling point | 798.5 °C (1,469.3 °F; 1,071.7 K) |
| Acidity (pKa) | 0.9, 3.8, 6.1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Adenosine monophosphate (AMP), also known as 5'-adenylic acid, is a nucleotide. AMP consists of a phosphate group, the sugar ribose, and the nucleobase adenine. It is an ester of phosphoric acid and the nucleoside adenosine.[1] As a substituent it takes the form of the prefix adenylyl-.[2]
AMP plays an important role in many cellular metabolic processes, being interconverted to ADP and/or ATP, as well as allosterically activating enzymes such as myophosphorylase-b. AMP is also a component in the synthesis of RNA.[3] AMP is present in all known forms of life.[4]
Production and degradation
AMP does not have the high energy phosphoanhydride bond associated with ADP and ATP. AMP can be produced from ADP by the myokinase (adenylate kinase) reaction when the ATP reservoir in the cell is low:[5][6]
- 2 ADP → ATP + AMP
Or AMP may be produced by the hydrolysis of one high energy phosphate bond of ADP:
- ADP + H2O → AMP + Pi
AMP can also be formed by hydrolysis of ATP into AMP and pyrophosphate:
- ATP + H2O → AMP + PPi
When RNA is broken down by living systems, nucleoside monophosphates, including adenosine monophosphate, are formed.
AMP can be regenerated to ATP as follows:
- AMP + ATP → 2 ADP (adenylate kinase in the opposite direction)
- ADP + Pi → ATP (this step is most often performed in aerobes by the ATP synthase during oxidative phosphorylation)
AMP can be converted into IMP by the enzyme myoadenylate deaminase, freeing an ammonia group.
In a catabolic pathway, the purine nucleotide cycle, adenosine monophosphate can be converted to uric acid, which is excreted from the body in mammals.[7]
Physiological role in regulation
AMP-activated kinase regulation
The eukaryotic cell enzyme 5' adenosine monophosphate-activated protein kinase, or AMPK, utilizes AMP for homeostatic energy processes during times of high cellular energy expenditure, such as exercise.[8] Since ATP cleavage, and corresponding phosphorylation reactions, are utilized in various processes throughout the body as a source of energy, ATP production is necessary to further create energy for those mammalian cells. AMPK, as a cellular energy sensor, is activated by decreasing levels of ATP, which is naturally accompanied by increasing levels of ADP and AMP.[9]
Though phosphorylation appears to be the main activator for AMPK, some studies suggest that AMP is an allosteric regulator as well as a direct agonist for AMPK.[10] Furthermore, other studies suggest that the high ratio of AMP:ATP levels in cells, rather than just AMP, activate AMPK.[11] For example, the AMP-activated kinases of Caenorhabditis elegans and Drosophila melanogaster were found to have been activated by AMP, while yeast and plant kinases were not allosterically activated by AMP.[11]
AMP binds to the γ-subunit of AMPK, leading to the activation of the kinase, and then eventually a cascade of other processes such as the activation of catabolic pathways and inhibition of anabolic pathways to regenerate ATP. Catabolic mechanisms, which generate ATP through the release of energy from breaking down molecules, are activated by the AMPK enzyme while anabolic mechanisms, which utilize energy from ATP to form products, are inhibited.[12] Though the γ-subunit can bind AMP/ADP/ATP, only the binding of AMP/ADP results in a conformational shift of the enzyme protein. This variance in AMP/ADP versus ATP binding leads to a shift in the dephosphorylation state for the enzyme.[13] The dephosphorylation of AMPK through various protein phosphatases completely inactivates catalytic function. AMP/ADP protects AMPK from being inactivated by binding to the γ-subunit and maintaining the dephosphorylation state.[14]
cAMP
AMP can also exist as a cyclic structure known as cyclic AMP (or cAMP). Within certain cells the enzyme adenylate cyclase makes cAMP from ATP, and typically this reaction is regulated by hormones such as adrenaline or glucagon. cAMP plays an important role in intracellular signaling.[15] In skeletal muscle, cyclic AMP, triggered by adrenaline, starts a cascade (cAMP-dependent pathway) for the conversion of myophosphorylase-b into the phosphorylated form of myophoshorylase-a for glycogenolysis.[16][17]
See also
References
- "Adenosine monophosphate (Compound)". PubChem. NCBI. Retrieved 30 April 2020.
- "Nomenclature of Carbohydrates: (Recommendations 1996)". Journal of Carbohydrate Chemistry. 16 (8): 1191–1280. 1997. doi:10.1080/07328309708005748.
- Jauker M, Griesser H, Richert C (November 2015). "Spontaneous Formation of RNA Strands, Peptidyl RNA, and Cofactors". Angewandte Chemie. 54 (48): 14564–9. doi:10.1002/anie.201506593. PMC 4678511. PMID 26435376.
- "Adenosine monophosphate". The Human Metabolome Database. Retrieved 3 July 2020.
- Baker, Julien S.; McCormick, Marie Clare; Robergs, Robert A. (2010). "Interaction among Skeletal Muscle Metabolic Energy Systems during Intense Exercise". Journal of Nutrition and Metabolism. 2010: 905612. doi:10.1155/2010/905612. ISSN 2090-0732. PMC 3005844. PMID 21188163.
- Valberg, Stephanie J. (1 January 2008), Kaneko, J. Jerry; Harvey, John W.; Bruss, Michael L. (eds.), "Chapter 15 - Skeletal Muscle Function", Clinical Biochemistry of Domestic Animals (Sixth Edition), San Diego: Academic Press, pp. 459–484, ISBN 978-0-12-370491-7, retrieved 10 October 2023
- Maiuolo J, Oppedisano F, Gratteri S, Muscoli C, Mollace V (June 2016). "Regulation of uric acid metabolism and excretion". International Journal of Cardiology. 213: 8–14. doi:10.1016/j.ijcard.2015.08.109. PMID 26316329.
- Richter EA, Ruderman NB (March 2009). "AMPK and the biochemistry of exercise: implications for human health and disease". The Biochemical Journal. 418 (2): 261–75. doi:10.1042/BJ20082055. PMC 2779044. PMID 19196246.
- Carling D, Mayer FV, Sanders MJ, Gamblin SJ (July 2011). "AMP-activated protein kinase: nature's energy sensor". Nature Chemical Biology. 7 (8): 512–8. doi:10.1038/nchembio.610. PMID 21769098.
- Faubert B, Vincent EE, Poffenberger MC, Jones RG (January 2015). "The AMP-activated protein kinase (AMPK) and cancer: many faces of a metabolic regulator". Cancer Letters. 356 (2 Pt A): 165–70. doi:10.1016/j.canlet.2014.01.018. PMID 24486219.
- Hardie DG (15 September 2011). "AMP-activated protein kinase—an energy sensor that regulates all aspects of cell function". Genes & Development. 25 (18): 1895–1908. doi:10.1101/gad.17420111. ISSN 0890-9369. PMC 3185962. PMID 21937710.
- Hardie DG (February 2011). "Energy sensing by the AMP-activated protein kinase and its effects on muscle metabolism". The Proceedings of the Nutrition Society. 70 (1): 92–9. doi:10.1017/S0029665110003915. PMID 21067629.
- Krishan S, Richardson DR, Sahni S (March 2015). "Adenosine monophosphate-activated kinase and its key role in catabolism: structure, regulation, biological activity, and pharmacological activation". Molecular Pharmacology. 87 (3): 363–77. doi:10.1124/mol.114.095810. PMID 25422142.
- Xiao B, Sanders MJ, Underwood E, Heath R, Mayer FV, Carmena D, Jing C, Walker PA, Eccleston JF, Haire LF, Saiu P, Howell SA, Aasland R, Martin SR, Carling D, Gamblin SJ (April 2011). "Structure of mammalian AMPK and its regulation by ADP". Nature. 472 (7342): 230–3. Bibcode:2011Natur.472..230X. doi:10.1038/nature09932. PMC 3078618. PMID 21399626.
- Ravnskjaer K, Madiraju A, Montminy M (2015). Metabolic Control. Handbook of Experimental Pharmacology. Vol. 233. Springer, Cham. pp. 29–49. doi:10.1007/164_2015_32. ISBN 9783319298047. PMID 26721678.
- Coffee, Carole J. (1999). Quick Look Medicine: Metabolism. Hayes Barton Press. ISBN 1-59377-192-4.
- "15.3: Glycogenolyis and its Regulation by Glucagon and Epinephrine Signaling". Biology LibreTexts. 1 January 2022. Retrieved 10 October 2023.
Further reading
- Ming D, Ninomiya Y, Margolskee RF (August 1999). "Blocking taste receptor activation of gustducin inhibits gustatory responses to bitter compounds". Proceedings of the National Academy of Sciences of the United States of America. 96 (17): 9903–8. Bibcode:1999PNAS...96.9903M. doi:10.1073/pnas.96.17.9903. PMC 22308. PMID 10449792.