Hepatitis D
Hepatitis D is a type of viral hepatitis[3] caused by the hepatitis delta virus (HDV).[4][5] HDV is one of five known hepatitis viruses: A, B, C, D, and E. HDV is considered to be a satellite (a type of subviral agent) because it can propagate only in the presence of the hepatitis B virus (HBV).[6] Transmission of HDV can occur either via simultaneous infection with HBV (coinfection) or superimposed on chronic hepatitis B or hepatitis B carrier state (superinfection).
| Hepatitis D | |
|---|---|
| Other names | Hepatitis delta |
| Specialty | Gastroenterology, infectious disease |
| Symptoms | Feeling tired, nausea and vomiting[1] |
| Complications | Cirrhosis[1] |
| Causes | Hepatitis D virus[1] |
| Diagnostic method | Immunoglobulin G[2] |
| Treatment | Antivirals, pegylated interferon alpha[2] |
| Medication | Bulevirtide |
HDV infecting a person with chronic hepatitis B (superinfection) is considered the most serious type of viral hepatitis due to its severity of complications.[7] These complications include a greater likelihood of experiencing liver failure in acute infections and a rapid progression to liver cirrhosis, with an increased risk of developing liver cancer in chronic infections.[8] In combination with hepatitis B virus, hepatitis D has the highest fatality rate of all the hepatitis infections, at 20%. A recent estimate from 2020 suggests that currently 48 million persons are infected with this virus.[9]
Virology
| Hepatitis delta virus | |
|---|---|
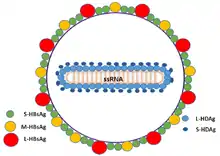 | |
| Schematic representation of the Hepatitis delta virus virion | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Ribozyviria |
| Family: | Kolmioviridae |
| Genus: | Deltavirus |
| Species[10] | |
| |
Structure and genome
The hepatitis delta viruses, or HDV, are eight species of negative-sense single-stranded RNA viruses (or virus-like particles) classified together as the genus Deltavirus, within the realm Ribozyviria.[11] The HDV virion is a small, spherical, enveloped particle with a 36 nm diameter; its viral envelope contains host phospholipids, as well as three proteins taken from the hepatitis B virus—the large, medium, and small hepatitis B surface antigens. This assembly surrounds an inner ribonucleoprotein (RNP) particle, which contains the genome surrounded by about 200 molecules of hepatitis D antigen (HDAg) for each genome. The central region of HDAg has been shown to bind RNA.[12] Several interactions are also mediated by a coiled-coil region at the N terminus of HDAg.[13][14]
The HDV genome is negative sense, single-stranded, closed circular RNA; with a genome of approximately 1700 nucleotides, HDV is the smallest "virus" known to infect animals. It has been proposed that HDV may have originated from a class of plant pathogens called viroids, which are much smaller than viruses.[15][16] Its genome is unique among animal viruses because of its high GC nucleotide content. Its nucleotide sequence is about 70% self-complementary, allowing the genome to form a partially double-stranded, rod-like RNA structure.[17] HDV strains are highly divergent; fusions of different strains exist and sequences had been deposited in public databases employing different start sites for the circular viral DNA involved. This had resulted in something of chaos with respect to molecular classification of this virus, a situation which has been resolved recently with the adoption of a proposed reference genome and a uniform classification system.[18]
Life cycle
Like hepatitis B, HDV gains entry into liver cells via the NTCP[19] bile transporter. HDV recognizes its receptor via the N-terminal domain of the large hepatitis B surface antigen, HBsAg.[20] Mapping by mutagenesis of this domain has shown that amino acid residues 9–15 make up the receptor-binding site.[21] After entering the hepatocyte, the virus is uncoated and the nucleocapsid translocated to the nucleus due to a signal in HDAg[22] Since the HDV genome does not code for an RNA polymerase to replicate the virus' genome, the virus makes use of the host cellular RNA polymerases. Initially thought to use just RNA polymerase II,[23][24] now RNA polymerases I and III have also been shown to be involved in HDV replication.[25] Normally RNA polymerase II utilizes DNA as a template and produces mRNA. Consequently, if HDV indeed utilizes RNA polymerase II during replication, it would be the only known animal pathogen capable of using a DNA-dependent polymerase as an RNA-dependent polymerase.
The RNA polymerases treat the RNA genome as double-stranded DNA due to the folded rod-like structure it is in. Three forms of RNA are made; circular genomic RNA, circular complementary antigenomic RNA, and a linear polyadenylated antigenomic RNA, which is the mRNA containing the open reading frame for the HDAg. Synthesis of antigenomic RNA occurs in the nucleolus, mediated by RNA polymerase I, whereas synthesis of genomic RNA takes place in the nucleoplasm, mediated by RNA polymerase II.[26] HDV RNA is synthesized first as linear RNA that contains many copies of the genome. The genomic and antigenomic RNA contain a sequence of 85 nucleotides, the hepatitis delta virus ribozyme, that acts as a ribozyme, which self-cleaves the linear RNA into monomers. These monomers are then ligated to form circular RNA.[27][28]
Delta antigens
| Hepatitis delta virus delta antigen | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 oligomerization domain of hepatitis delta antigen | |||||||||
| Identifiers | |||||||||
| Symbol | HDV_ag | ||||||||
| Pfam | PF01517 | ||||||||
| InterPro | IPR002506 | ||||||||
| SCOP2 | 1a92 / SCOPe / SUPFAM | ||||||||
| |||||||||
A significant difference between viroids and HDV is that, while viroids produce no proteins, HDV is known to produce one protein, namely HDAg. It comes in two forms; a 27kDa large-HDAg, and a small-HDAg of 24kDa. The N-terminals of the two forms are identical, they differ by 19 more amino acids in the C-terminal of the large HDAg.[29] Both isoforms are produced from the same reading frame which contains an UAG stop codon at codon 196, which normally produces only the small-HDAg. However, editing by cellular enzyme adenosine deaminase-1 changes the stop codon to UGG, allowing the large-HDAg to be produced.[29][30] Despite having 90% identical sequences, these two proteins play diverging roles during the course of an infection. HDAg-S is produced in the early stages of an infection and enters the nucleus and supports viral replication. HDAg-L, in contrast, is produced during the later stages of an infection, acts as an inhibitor of viral replication, and is required for assembly of viral particles.[31][32][33] Thus RNA editing by the cellular enzymes is critical to the virus' life cycle because it regulates the balance between viral replication and virion assembly.
Antigenic loop infectivity
The HDV envelope protein has three of the HBV surface proteins anchored to it. The S region of the genome is most commonly expressed and its main function is to assemble subviral particles. HDV antigen proteins combine with the viral genome to form a ribonucleoprotein (RNP) which when enveloped with the subviral particles can form viral-like particles that are almost identical to mature HDV, but they are not infectious. Researchers had concluded that the determinant of infectivity of HDV was within the N-terminal pre-S1 domain of the large protein (L). It was found to be a mediator in binding to the cellular receptor. Researchers Georges Abou Jaoudé and Camille Sureau published an article in 2005 that studied the role of the antigenic loop, found in HDV envelope proteins, in the infectivity of the virus. The antigenic loop, like the N-terminal pre-S1 domain of the large protein, is exposed at the virion surface. Jaoudé and Sureau's study provided evidence that the antigenic loop may be an important factor in HDV entry into the host cell and by mutating parts of the antigenic loop, the infectivity of HDV may be minimized.[34]
Transmission
The routes of transmission of hepatitis D are similar to those for hepatitis B. Infection is largely restricted to persons at high risk of hepatitis B infection, particularly injecting drug users and persons receiving clotting factor concentrates. Worldwide more than 15 million people are co-infected. HDV is rare in most developed countries, and is mostly associated with intravenous drug use. However, HDV is much more common in the immediate Mediterranean region, sub-Saharan Africa, the Middle East, and the northern part of South America.[35] In all, about 20 million people may be infected with HDV.[36]
Prevention
The vaccine for hepatitis B protects against hepatitis D virus because of the latter's dependence on the presence of hepatitis B virus for it to replicate.[37][38]
In absence of a specific vaccine against delta virus, the vaccine against HBV must be given soon after birth in risk groups.
Treatment
Hepatitis D is generally considered the dominant virus over hepatitis B except in rare instances. Current established treatments for chronic hepatitis D include conventional or pegylated interferon alpha therapy.[39] Latest evidence suggests that pegylated interferon alpha is effective in reducing the viral load and the effect of the disease during the time the drug is given, but the benefit generally stops if the drug is discontinued.[40] The efficiency of this treatment does not usually exceed ~20%, and late relapse after therapy has been reported.[41][42]
In May 2020, the Committee for Medicinal Products for Human Use of the European Medicines Agency approved the antiviral Hepcludex (bulevirtide) to treat hepatitis D and B. Bulevirtide binds and inactivates the sodium/bile acid cotransporter, blocking both viruses from entering hepatocytes.[43][44]
Epidemiology
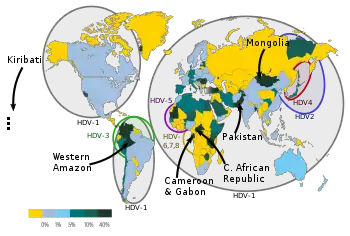
Those affected are individuals who have been infected with Hepatitis B virus as the Hepatitis D (HDV) virus needs the HBsAg (hepatitis B surface antigen) for packaging and transmission. The disease is present worldwide. Infection with HDV is a major medical scourge in low income regions of the globe in which the HBV remains endemic.[46] It is therefore most prevalent in countries where HBV infection is also common, currently the Amazon basin and low income regions of Asia and Africa. Improved measures to control HBV in industrialised countries (such as by vaccination) have also reduced the prevalence of HDV, with the main remaining at-risk populations in those countries being injection drug users and immigrants from endemic HDV areas.[46]
History
Hepatitis D virus was first reported in 1977 as a nuclear antigen in patients infected with HBV who had severe liver disease.[47] This nuclear antigen was then thought to be a hepatitis B antigen and was called the delta antigen. Subsequent experiments in chimpanzees showed that the hepatitis delta antigen (HDAg) was a structural part of a pathogen that required HBV infection to produce a complete viral particle.[48] The entire genome was cloned and sequenced in 1986. It was subsequently placed in its own genus: Deltavirus.[49][50]
Lábrea fever
| Lábrea fever | |
|---|---|
| Other names | Lábrea's black fever, Lábrea hepatitis, Santa Marta fever |
| Specialty | Infectious disease |
| Usual onset | sudden |
| Duration | approx. 1 week |
| Prevention | HBV vaccination |
| Prognosis | death |
Lábrea fever is a lethal tropical infection discovered in the 1950s in the city of Lábrea, in the Brazilian Amazon basin, where it occurs mostly in the area south of the Amazon River, in the states of Acre, Amazonas, and Rondônia. The disease has also been diagnosed in Colombia and Peru. It is now known to be a coinfection or superinfection of hepatitis B (HBV) with hepatitis D.[51]
Lábrea fever has a sudden onset, with jaundice, anorexia (lack of appetite), hematemesis (vomiting of blood), headache, fever and severe prostration. Death occurs by acute liver failure (ALF). In the last phase, neurological symptoms such as agitation, delirium, convulsions and hemorrhagic coma commonly appear. These symptoms arise from a fulminant hepatitis which may kill in less than a week, and which characteristically affects children and young adults, and more males than females. It is accompanied also by an encephalitis in many cases. The disease is highly lethal: in a study carried out in 1986 at Boca do Acre, also in the Amazon, 39 patients out of 44 died in the acute phase of the disease.[51] Survivors may develop chronic disease.
The main discovery of delta virus and HBV association was done by Gilberta Bensabath, of the Instituto Evandro Chagas, of Belém, state of Pará, and her collaborators.
Infected patients show extensive destruction of liver tissue, with steatosis of a particular type (microsteatosis, characterized by small fat droplets inside the cells), and infiltration of large numbers of inflammatory cells called morula cells, comprised mainly by macrophages containing delta virus antigens.
In the 1987 Boca do Acre study, scientists did an epidemiological survey and reported delta virus infection in 24% of asymptomatic HBV carriers, 29% of acute nonfulminant hepatitis B cases, 74% of fulminant hepatitis B cases, and 100% of chronic hepatitis B cases.[51] The delta virus seems to be endemic in the Amazon region.
Evolution
Three genotypes (I–III) were originally described. Genotype I has been isolated in Europe, North America, Africa and some Asia. Genotype II has been found in Japan, Taiwan, and Yakutia (Russia). Genotype III has been found exclusively in South America (Peru, Colombia, and Venezuela). Some genomes from Taiwan and the Okinawa islands have been difficult to type but have been placed in genotype 2. However it is now known that there are at least 8 genotypes of this virus (HDV-1 to HDV-8).[52] Phylogenetic studies suggest an African origin for this pathogen.[35]
An analysis of 36 strains of genotype 3 estimated that the most recent common ancestor of these strains originated around 1930.[53] This genotype spread exponentially from early 1950s to the 1970s in South America. The substitution rate was estimated to be 1.07×10−3 substitutions per site per year. Another study[54] found an overall evolution rate of 3.18×10−3 substitutions per site per year. The mutation rate varied with position : the hypervariable region evolved faster (4.55×10−3 substitutions per site per year) than the hepatitis delta antigen coding region (2.60×10−3 substitutions per site per year) and the autocatalytic region (1.11×10−3 substitutions per site per year). A third study suggested a mutation rate between 9.5×10−3 to 1.2×10−3 substitutions/site/year.[55]
Genotypes, with the exception of type 1, appear to be restricted to certain geographical areas: HDV-2 (previously HDV-IIa) is found in Japan, Taiwan and Yakutia; HDV-4 (previously HDV-IIb) in Japan and Taiwan; HDV-3 in the Amazonian region; HDV-5, HDV-6, HDV-7 and HDV-8 in Africa.[56] Genotype 8 has also been isolated from South America. This genotype is usually only found in Africa and may have been imported into South America during the slave trade.[57]
HDV-specific CD8+ T cells can control the virus, but it has been found HDV mutates to escape detection by CD8+ T cells.[58]
Related species
A few other viruses with similarity to HDV have been described in species other than humans. Unlike HDV, none of them depend on a Hepadnaviridae (HBV family) virus to replicate. These agents have rod-like structure, a delta antigen, and a ribozyme.[59] HDV and all such relatives are classified in their own realm, Ribozyviria, by the International Committee on Taxonomy of Viruses.[11]
References
- "Hepatitis D | NIDDK". National Institute of Diabetes and Digestive and Kidney Diseases. Retrieved 10 September 2019.
- "Hepatitis D". www.who.int. Retrieved 10 September 2019.
- "Hepatitis (Viral) NIDDK". The National Institute of Diabetes and Digestive and Kidney Diseases. Retrieved 2020-06-19.
- Farci P (2003). "Delta hepatitis: an update". Journal of Hepatology. 39 (Suppl 1): S212–9. doi:10.1016/s0168-8278(03)00331-3. PMID 14708706.
- Magnius L, Taylor J, Mason WS, Sureau C, Dény P, Norder H (December 2018). "ICTV Virus Taxonomy Profile: Deltavirus". The Journal of General Virology. 99 (12): 1565–1566. doi:10.1099/jgv.0.001150. PMID 30311870.
- Makino S, Chang MF, Shieh CK, Kamahora T, Vannier DM, Govindarajan S, Lai MM (1987). "Molecular cloning and sequencing of a human hepatitis delta (delta) virus RNA". Nature. 329 (6137): 343–6. Bibcode:1987Natur.329..343M. doi:10.1038/329343a0. PMID 3627276. S2CID 4368061.
- "Hepatitis D". www.who.int. Retrieved 2020-09-20.
- Fattovich G, Giustina G, Christensen E, Pantalena M, Zagni I, Realdi G, Schalm SW (March 2000). "Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep)". Gut. 46 (3): 420–6. doi:10.1136/gut.46.3.420. PMC 1727859. PMID 10673308.
- Miao Z, Zhang S, Ou X, Li S, Ma Z, Wang W, Peppelenbosch MP, Liu J, Pan Q (April 2020). "Estimating the Global Prevalence, Disease Progression, and Clinical Outcome of Hepatitis Delta Virus Infection". The Journal of Infectious Diseases. 221 (10): 1677–1687. doi:10.1093/infdis/jiz633. PMC 7184909. PMID 31778167.
- "ICTV 9th Report (2011) Deltavirus". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 30 January 2019. Retrieved 30 January 2019.
- "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Retrieved 5 August 2021.
- Poisson F, Roingeard P, Baillou A, Dubois F, Bonelli F, Calogero RA, Goudeau A (November 1993). "Characterization of RNA-binding domains of hepatitis delta antigen". The Journal of General Virology. 74 (Pt 11): 2473–8. doi:10.1099/0022-1317-74-11-2473. PMID 8245865.
- Zuccola HJ, Rozzelle JE, Lemon SM, Erickson BW, Hogle JM (July 1998). "Structural basis of the oligomerization of hepatitis delta antigen". Structure. 6 (7): 821–30. doi:10.1016/S0969-2126(98)00084-7. PMID 9687364.
- This article incorporates text from the public domain Pfam and InterPro: IPR002506
- Elena SF, Dopazo J, Flores R, Diener TO, Moya A (July 1991). "Phylogeny of viroids, viroidlike satellite RNAs, and the viroidlike domain of hepatitis delta virus RNA". Proceedings of the National Academy of Sciences of the United States of America. 88 (13): 5631–4. Bibcode:1991PNAS...88.5631E. doi:10.1073/pnas.88.13.5631. PMC 51931. PMID 1712103.
- Sureau C (2006). "The role of the HBV envelope proteins in the HDV replication cycle". Hepatitis Delta Virus. Current Topics in Microbiology and Immunology. Vol. 307. pp. 113–31. doi:10.1007/3-540-29802-9_6. ISBN 978-3-540-29801-4. PMID 16903223.
- Saldanha JA, Thomas HC, Monjardino JP (July 1990). "Cloning and sequencing of RNA of hepatitis delta virus isolated from human serum". The Journal of General Virology. 71 (7): 1603–6. doi:10.1099/0022-1317-71-7-1603. PMID 2374010.
- Miao Z, Zhang S, Ma Z, Hakim MS, Wang W, Peppelenbosch MP, Pan Q (January 2019). "Recombinant identification, molecular classification and proposed reference genomes for hepatitis delta virus". Journal of Viral Hepatitis. 26 (1): 183–190. doi:10.1111/jvh.13010. PMC 7379554. PMID 30260538.
- Yan H, Zhong G, Xu G, He W, Jing Z, Gao Z, Huang Y, Qi Y, Peng B, Wang H, Fu L, Song M, Chen P, Gao W, Ren B, Sun Y, Cai T, Feng X, Sui J, Li W (November 2012). "Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus". eLife. 1: e00049. doi:10.7554/eLife.00049. PMC 3485615. PMID 23150796.
- Engelke M, Mills K, Seitz S, Simon P, Gripon P, Schnölzer M, Urban S (April 2006). "Characterization of a hepatitis B and hepatitis delta virus receptor binding site". Hepatology. 43 (4): 750–60. doi:10.1002/hep.21112. PMID 16557545. S2CID 23549907.
- Schulze A, Schieck A, Ni Y, Mier W, Urban S (February 2010). "Fine mapping of pre-S sequence requirements for hepatitis B virus large envelope protein-mediated receptor interaction". Journal of Virology. 84 (4): 1989–2000. doi:10.1128/JVI.01902-09. PMC 2812397. PMID 20007265.
- Xia YP, Yeh CT, Ou JH, Lai MM (February 1992). "Characterization of nuclear targeting signal of hepatitis delta antigen: nuclear transport as a protein complex". Journal of Virology. 66 (2): 914–21. doi:10.1128/JVI.66.2.914-921.1992. PMC 240792. PMID 1731113.
- Lehmann E, Brueckner F, Cramer P (November 2007). "Molecular basis of RNA-dependent RNA polymerase II activity". Nature. 450 (7168): 445–9. Bibcode:2007Natur.450..445L. doi:10.1038/nature06290. hdl:11858/00-001M-0000-0015-7EE1-9. PMID 18004386. S2CID 4393153.
- Filipovska J, Konarska MM (January 2000). "Specific HDV RNA-templated transcription by pol II in vitro". RNA. 6 (1): 41–54. doi:10.1017/S1355838200991167. PMC 1369892. PMID 10668797.
- Greco-Stewart VS, Schissel E, Pelchat M (March 2009). "The hepatitis delta virus RNA genome interacts with the human RNA polymerases I and III". Virology. 386 (1): 12–5. doi:10.1016/j.virol.2009.02.007. PMID 19246067.
- Li YJ, Macnaughton T, Gao L, Lai MM (July 2006). "RNA-templated replication of hepatitis delta virus: genomic and antigenomic RNAs associate with different nuclear bodies". Journal of Virology. 80 (13): 6478–86. doi:10.1128/JVI.02650-05. PMC 1488965. PMID 16775335.
- Branch AD, Benenfeld BJ, Baroudy BM, Wells FV, Gerin JL, Robertson HD (February 1989). "An ultraviolet-sensitive RNA structural element in a viroid-like domain of the hepatitis delta virus". Science. 243 (4891): 649–52. Bibcode:1989Sci...243..649B. doi:10.1126/science.2492676. PMID 2492676.
- Wu HN, Lin YJ, Lin FP, Makino S, Chang MF, Lai MM (March 1989). "Human hepatitis delta virus RNA subfragments contain an autocleavage activity". Proceedings of the National Academy of Sciences of the United States of America. 86 (6): 1831–5. Bibcode:1989PNAS...86.1831W. doi:10.1073/pnas.86.6.1831. PMC 286798. PMID 2648383.
- Weiner AJ, Choo QL, Wang KS, Govindarajan S, Redeker AG, Gerin JL, Houghton M (February 1988). "A single antigenomic open reading frame of the hepatitis delta virus encodes the epitope(s) of both hepatitis delta antigen polypeptides p24 delta and p27 delta". Journal of Virology. 62 (2): 594–9. doi:10.1128/JVI.62.2.594-599.1988. PMC 250573. PMID 2447291.
- Jayan GC, Casey JL (December 2002). "Inhibition of hepatitis delta virus RNA editing by short inhibitory RNA-mediated knockdown of ADAR1 but not ADAR2 expression". Journal of Virology. 76 (23): 12399–404. doi:10.1128/JVI.76.23.12399-12404.2002. PMC 136899. PMID 12414985.
- Sato S, Cornillez-Ty C, Lazinski DW (August 2004). "By inhibiting replication, the large hepatitis delta antigen can indirectly regulate amber/W editing and its own expression". Journal of Virology. 78 (15): 8120–34. doi:10.1128/JVI.78.15.8120-8134.2004. PMC 446097. PMID 15254184.
- Taylor JM (2006). "Structure and replication of hepatitis delta virus RNA". Hepatitis Delta Virus. Current Topics in Microbiology and Immunology. Vol. 307. pp. 1–23. doi:10.1007/3-540-29802-9_1. ISBN 978-3-540-29801-4. PMID 16903218.
- Chang MF, Chen CJ, Chang SC (February 1994). "Mutational analysis of delta antigen: effect on assembly and replication of hepatitis delta virus". Journal of Virology. 68 (2): 646–53. doi:10.1128/JVI.68.2.646-653.1994. PMC 236498. PMID 8289368.
- Jaoudé GA, Sureau C (August 2005). "Role of the antigenic loop of the hepatitis B virus envelope proteins in infectivity of hepatitis delta virus". Journal of Virology. 79 (16): 10460–6. CiteSeerX 10.1.1.570.4147. doi:10.1128/jvi.79.16.10460-10466.2005. PMC 1182656. PMID 16051838.
- Radjef N, Gordien E, Ivaniushina V, Gault E, Anaïs P, Drugan T, Trinchet JC, Roulot D, Tamby M, Milinkovitch MC, Dény P (March 2004). "Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades". Journal of Virology. 78 (5): 2537–44. doi:10.1128/JVI.78.5.2537-2544.2004. PMC 369207. PMID 14963156.
- Taylor JM (January 2006). "Hepatitis delta virus". Virology. 344 (1): 71–6. doi:10.1016/j.virol.2005.09.033. PMID 16364738.
- "U.S. National Library of Medicine "Delta Agent (hepatitis D)"".
- Tayor JM (2009). Desk Encyclopedia of Human and Medical Virology. Boston: Academic Press. p. 121. ISBN 978-0-12-375147-8.
- Yurdaydin C, Idilman R (August 2015). "Therapy of Delta Hepatitis". Cold Spring Harbor Perspectives in Medicine. 5 (10): a021543. doi:10.1101/cshperspect.a021543. PMC 4588130. PMID 26253093.
- Abbas Z, Khan MA, Salih M, Jafri W (December 2011). Abbas Z (ed.). "Interferon alpha for chronic hepatitis D". The Cochrane Database of Systematic Reviews (12): CD006002. doi:10.1002/14651858.CD006002.pub2. PMC 6823236. PMID 22161394.
- Heidrich B, Yurdaydın C, Kabaçam G, Ratsch BA, Zachou K, Bremer B, Dalekos GN, Erhardt A, Tabak F, Yalcin K, Gürel S, Zeuzem S, Cornberg M, Bock CT, Manns MP, Wedemeyer H (July 2014). "Late HDV RNA relapse after peginterferon alpha-based therapy of chronic hepatitis delta". Hepatology. 60 (1): 87–97. doi:10.1002/hep.27102. PMID 24585488. S2CID 205892640.
- Pascarella S, Negro F (January 2011). "Hepatitis D virus: an update". Liver International. 31 (1): 7–21. doi:10.1111/j.1478-3231.2010.02320.x. PMID 20880077. S2CID 29142477.
- Francisco EM (2020-05-29). "Hepcludex". European Medicines Agency. Archived from the original on 2020-06-15. Retrieved 2020-08-06.
- "Bulevirtide - MYR Pharma - AdisInsight". adisinsight.springer.com. Retrieved 2020-08-06.
MYR Pharmaceuticals receives Conditional Marketing Authorisation by the European Commission for bulevirtide in the European Union for Hepatitis B and D
- Rizzetto M (2020). "Epidemiology of the Hepatitis D virus". WikiJournal of Medicine. 7: 7. doi:10.15347/wjm/2020.001.
- Rizzetto M (July 2015). "Hepatitis D Virus: Introduction and Epidemiology". Cold Spring Harbor Perspectives in Medicine. 5 (7): a021576. doi:10.1101/cshperspect.a021576. PMC 4484953. PMID 26134842.
- Rizzetto M, Canese MG, Aricò S, Crivelli O, Trepo C, Bonino F, Verme G (December 1977). "Immunofluorescence detection of new antigen-antibody system (delta/anti-delta) associated to hepatitis B virus in liver and in serum of HBsAg carriers". Gut. 18 (12): 997–1003. doi:10.1136/gut.18.12.997. PMC 1411847. PMID 75123.
- Rizzetto M, Canese MG, Purcell RH, London WT, Sly LD, Gerin JL (Nov–Dec 1981). "Experimental HBV and delta infections of chimpanzees: occurrence and significance of intrahepatic immune complexes of HBcAg and delta antigen". Hepatology. 1 (6): 567–74. doi:10.1002/hep.1840010602. PMID 7030907. S2CID 83892580.
- Wang KS, Choo QL, Weiner AJ, Ou JH, Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL, Houghton M (Oct 9–15, 1986). "Structure, sequence and expression of the hepatitis delta (delta) viral genome". Nature. 323 (6088): 508–14. Bibcode:1986Natur.323..508W. doi:10.1038/323508a0. PMID 3762705. S2CID 4265339.
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (2005). "Deltavirus". Eight Report of the International Committee on Taxonomy of Viruses. London: 735–8.
- Bensabath G, Hadler SC, Soares MC, Fields H, Dias LB, Popper H, Maynard JE (1987). "Hepatitis delta virus infection and Labrea hepatitis. Prevalence and role in fulminant hepatitis in the Amazon Basin". JAMA. 258 (4): 479–83. doi:10.1001/jama.1987.03400040077025. PMID 3599343.
- Celik I, Karatayli E, Cevik E, Kabakçi SG, Karatayli SC, Dinç B, et al. (December 2011). "Complete genome sequences and phylogenetic analysis of hepatitis delta viruses isolated from nine Turkish patients". Archives of Virology. 156 (12): 2215–20. doi:10.1007/s00705-011-1120-y. PMID 21984217. S2CID 31356.
- Alvarado-Mora MV, Romano CM, Gomes-Gouvêa MS, Gutierrez MF, Carrilho FJ, Pinho JR (August 2011). "Dynamics of hepatitis D (delta) virus genotype 3 in the Amazon region of South America". Infection, Genetics and Evolution. 11 (6): 1462–8. doi:10.1016/j.meegid.2011.05.020. PMID 21645647.
- Chao YC, Tang HS, Hsu CT (August 1994). "Evolution rate of hepatitis delta virus RNA isolated in Taiwan". Journal of Medical Virology. 43 (4): 397–403. doi:10.1002/jmv.1890430414. PMID 7964650. S2CID 22539505.
- Homs M, Rodriguez-Frias F, Gregori J, Ruiz A, Reimundo P, Casillas R, Tabernero D, Godoy C, Barakat S, Quer J, Riveiro-Barciela M, Roggendorf M, Esteban R, Buti M (2016). "Evidence of an Exponential Decay Pattern of the Hepatitis Delta Virus Evolution Rate and Fluctuations in Quasispecies Complexity in Long-Term Studies of Chronic Delta Infection". PLOS ONE. 11 (6): e0158557. Bibcode:2016PLoSO..1158557H. doi:10.1371/journal.pone.0158557. PMC 4928832. PMID 27362848.
- Le Gal F, Gault E, Ripault MP, Serpaggi J, Trinchet JC, Gordien E, Dény P (September 2006). "Eighth major clade for hepatitis delta virus". Emerging Infectious Diseases. 12 (9): 1447–50. doi:10.3201/eid1209.060112. PMC 3294742. PMID 17073101.
- Barros LM, Gomes-Gouvêa MS, Pinho JR, Alvarado-Mora MV, Dos Santos A, Mendes-Corrêa MC, Caldas AJ, Sousa MT, Santos MD, Ferreira AS (September 2011). "Hepatitis Delta virus genotype 8 infection in Northeast Brazil: inheritance from African slaves?". Virus Research. 160 (1–2): 333–9. doi:10.1016/j.virusres.2011.07.006. PMID 21798297.
- Karimzadeh H, Kiraithe MM, Oberhardt V, Salimi Alizei E, Bockmann J, Schulze Zur Wiesch J, et al. (May 2019). "Mutations in Hepatitis D Virus Allow It to Escape Detection by CD8+ T Cells and Evolve at the Population Level". Gastroenterology. 156 (6): 1820–1833. doi:10.1053/j.gastro.2019.02.003. PMC 6486497. PMID 30768983.
- Paraskevopoulou S, Pirzer F, Goldmann N, Schmid J, Corman VM, Gottula LT, et al. (July 2020). "Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus". Proceedings of the National Academy of Sciences of the United States of America. 117 (30): 17977–17983. doi:10.1073/pnas.2006750117. PMC 7395443. PMID 32651267.
Bibliography
- Specter SC, ed. (1999). Viral Hepatitis: Diagnosis, Therapy, and Prevention. Humana Press. ISBN 0-89603-424-0.
- da Fonseca JC (2004). "[Hepatitis fulminant in Brazilian Amazon]". Revista da Sociedade Brasileira de Medicina Tropical. 37. 37 Suppl 2: 93–5. doi:10.1590/s0037-86822004000700015. PMID 15586904.
- Bensabath G, Soares M (2004). "[The evolution of knowledge about viral hepatitis in Amazon region: from epidemiology and etiology to the prophilaxy]". Revista da Sociedade Brasileira de Medicina Tropical. 37 () Suppl 2): 14–26. doi:10.1590/S0037-86822004000700003. PMID 15586892.
- Fonseca JC, Souza RA, Brasil LM, Araújo JR, Ferreira LC (2004). "Fulminant hepatic failure in children and adolescents in Northern Brazil". Revista da Sociedade Brasileira de Medicina Tropical. 37 (1): 67–9. doi:10.1590/S0037-86822004000100019. PMID 15042190.
External links
- World Health Organization Fact sheet on Hepatitis D
- ICTV Report: Deltavirus
- Viralzone: Deltavirus
- "Hepatitis delta virus". NCBI Taxonomy Browser. 12475.