Ergocryptine
 | |
| Identifiers | |
|---|---|
| CAS Number | |
| PubChem CID | |
| IUPHAR/BPS | |
| ChemSpider | |
| UNII | |
| ChEMBL | |
| CompTox Dashboard (EPA) | |
| ECHA InfoCard | 100.007.384 |
| Chemical and physical data | |
| Formula | C32H41N5O5 |
| Molar mass | 575.710 g·mol−1 |
| 3D model (JSmol) | |
SMILES
| |
InChI
| |
| | |
Ergocryptine is an ergopeptine and one of the ergot alkaloids. It is isolated from ergot or fermentation broth and it serves as starting material for the production of bromocriptine.[1]
Chemistry
Ergocryptine is a mixture of two very similar compounds, alpha- and beta-ergocryptine.[2] The beta differs from the alpha form only in the position of a single methyl group, which is a consequence of the biosynthesis in which the proteinogenic amino acid leucine is replaced by isoleucine.
Biosynthesis
The biosynthetic pathways to ergocryptine starts with the prenylation of L-tryptophan in an SN1 fashion with Dimethylallylpyrophosphate (DMAPP). The DMAPP is derived from Mevalonic Acid. This reaction is catalyzed by a prenyltransferase enzyme (Prenyltransferase 4-dimethylallyltryptophan synthase) named FgaPT2 in A. fumigatus.[3][4] An X-ray structure of the prenyltransferase FgaPT2 and tryptophan has been reported, and used to propose a three step mechanism (formation of allylic carbocation, nucleophilic attack of tryptophan on the carbocation, then deprotonation to restore aromaticity and generate the product, 4-dimethylallyltryptophan (DMAT)).[4] DMAT is then N-methylated at the amino of the tryptophan backbone with the EasF enzyme, named FgaMT in A. fumigatus. S-adenosylmethionine (SAM) being the methyl source.[5]
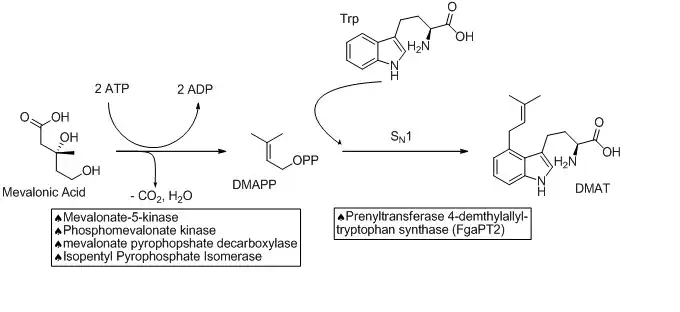
The next step in the biosynthesis of ergocryptine is the transformation of 4-dimethylallyl abrine to Chanoclavine-I. It has been shown that the enzymes EasE and EasC (FgaOx1 and FgaCat in A. fumigatus, respectively) are both required to generate Chanoclavine-I from 4-DMA abrine.[6] Mutation experiments altering these enzymes independently stopped the pathway at abrine. This indicates that cooperation between EasE and EasC is necessary.

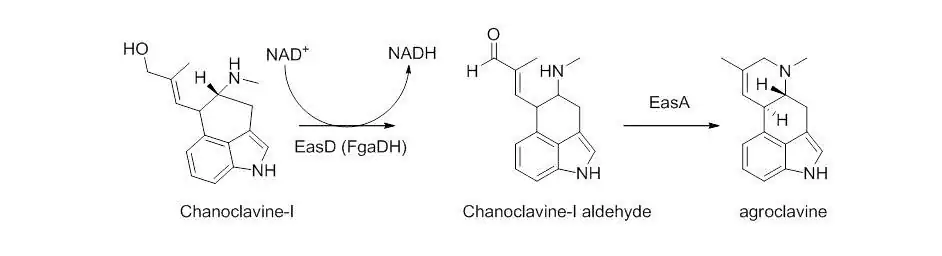
Chanocalvine-I is then oxidized to chanoclavine-I aldehyde with NAD+ dependent enzyme EasD (FgaDH in A. fumigatus). Chanoclavine-I aldehyde is a branch point, leading to different ergot alkaloids, depending on the specific fungus. In C. purpurea, chanoclavine-I aldehyde is converted to argoclavine with EasA, referred to as the old yellow enzyme or FgaOx3. This process occurs via keto-enol tautomerization to facilitate rotation about a carbon-carbon bond, followed by tautomerization back to the aldehyde, and condensation with the proximal secondary amine.[4][7] The iminium species created by cyclization is then reduced to the tertiary amine, yielding agroclavine.

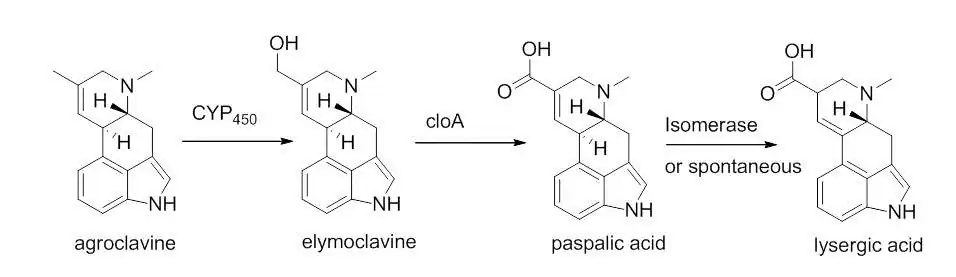
A cyctochrome P-450 monooxygenase enzyme catalyzes a two electron oxidation of agroclavne to the corresponding primary alcohol, elymoclavine.[8] Elymoclavine is then oxidized by four electrons by a P450 monooxygenase to give paspalic acid. Paspalic acid then undergoes isomerization of the carbon-carbon double bond that is in conjugation with the acid, to give D-lysergic acid.
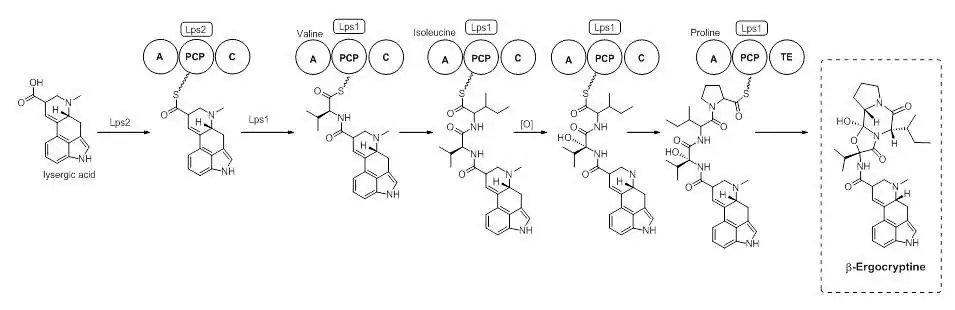
Lysergic Acid is a branch point in the biosynthesis of ergoamides and ergopeptines. On the path to ergocryptine, an ergopeptine, the tripeptide is installed by a Non-Ribosomal Peptide Synthase (NRPS). It has been shown that there are two enzymes, D-lysergyl peptide synthases (LPS) 1 and 2, which are responsible for the tripeptide connection to lysergic acid.[9] The timing of the oxidation of valine to an alcohol is not exactly known. However, it is speculated that the oxidation occurs while bound to the NRPS LPS2.[10] Ergocryptine is found in two forms, differing in the amino acid used by the NRPS. The alpha form contains the amino acid leucine, while the beta-form uses the amino acid isoleucine.[4]
See also
References
- ↑ Kren V, Cvak L (1999). Ergot: the genus Claviceps. Amsterdam: Harwood Academic Publishers. pp. 399–401.
- ↑ Yates SG, Plattner RD, Garner GB (1985). "Detection of ergopeptine alkaloids in endophyte-infected, toxic Ky-31 tall fescue by mass spectrometry/mass spectrometry" (PDF). Journal of Agricultural and Food Chemistry. 33 (4): 719–722. doi:10.1021/jf00064a038.
- ↑ Lee SL, Floss HG, Heinstein P (November 1976). "Purification and properties of dimethylallylpyrophosphate:tryptopharm dimethylallyl transferase, the first enzyme of ergot alkaloid biosynthesis in Claviceps. sp. SD 58". Archives of Biochemistry and Biophysics. 177 (1): 84–94. doi:10.1016/0003-9861(76)90418-5. PMID 999297.
- 1 2 3 4 Gerhards N, Neubauer L, Tudzynski P, Li SM (December 2014). "Biosynthetic pathways of ergot alkaloids". Toxins. 6 (12): 3281–95. doi:10.3390/toxins6123281. PMC 4280535. PMID 25513893.
- ↑ Rigbers O, Li SM (October 2008). "Ergot alkaloid biosynthesis in Aspergillus fumigatus. Overproduction and biochemical characterization of a 4-dimethylallyltryptophan N-methyltransferase". The Journal of Biological Chemistry. 283 (40): 26859–68. doi:10.1074/jbc.M804979200. PMID 18678866.
- ↑ Goetz KE, Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG (June 2011). "Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus". Current Genetics. 57 (3): 201–11. doi:10.1007/s00294-011-0336-4. PMID 21409592. S2CID 3031547.
- ↑ Coyle CM, Cheng JZ, O'Connor SE, Panaccione DG (June 2010). "An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways". Applied and Environmental Microbiology. 76 (12): 3898–903. doi:10.1128/AEM.02914-09. PMC 2893504. PMID 20435769.
- ↑ Haarmann T, Machado C, Lübbe Y, Correia T, Schardl CL, Panaccione DG, Tudzynski P (June 2005). "The ergot alkaloid gene cluster in Claviceps purpurea: extension of the cluster sequence and intra species evolution". Phytochemistry. 66 (11): 1312–20. doi:10.1016/j.phytochem.2005.04.011. PMID 15904941.
- ↑ Walzel B, Riederer B, Keller U (March 1997). "Mechanism of alkaloid cyclopeptide synthesis in the ergot fungus Claviceps purpurea". Chemistry & Biology. 4 (3): 223–30. doi:10.1016/s1074-5521(97)90292-1. PMID 9115414.
- ↑ Keller U (1995). Genetics and Biochemistry of Antibiotic Production. Boston: Butterworth-Heinemann.