TOX
Thymocyte selection-associated high mobility group box protein TOX is a protein that in humans is encoded by the TOX gene.[5][6][7] TOX drives T-cell exhaustion[8][9] and plays a role in innate lymphoid cell development.[10][11]
| TOX | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | TOX, TOX1, thymocyte selection associated high mobility group box | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 606863 MGI: 2181659 HomoloGene: 8822 GeneCards: TOX | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
Structure
The TOX gene encodes a protein that belongs to a large superfamily of chromatin associated proteins that share an approximately 75 amino acid DNA binding motif, the HMG (high mobility group)-box (named after that found in the canonical member of the family, high mobility group protein 1). Some high mobility group (HMG) box proteins (e.g., LEF1) contain a single HMG box motif and bind DNA in a sequence-specific manner, while other members of this family (e.g., HMGB1) have multiple HMG boxes and bind DNA in a sequence-independent but structure-dependent manner. While TOX has a single HMG-box motif,[7] it is predicted to bind DNA in a sequence-independent manner.[12]
TOX subfamily
TOX is a member of a small subfamily of proteins (TOX2, TOX3, and TOX4) that share almost identical HMG-box sequences.[12] TOX2 has been identified to play a role in the differentiation of T follicular helper cell.[13] TOX2 is thought to be a downstream signal of BCL-6.[13] TOX3 has been identified as a breast cancer susceptibility locus.[14][15] TOX is highly expressed in the thymus, the site of development of T lymphocytes.[10] Knockout mice that lack TOX have a severe defect in development of certain subsets of T lymphocytes.[16]
Function
T cell exhaustion
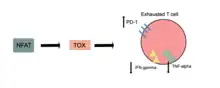
TOX is necessary for T cell persistence but also drives T cell exhaustion.[17][18][19] An increase in TOX expression is characterized by a weakening of the effector functions of the cytotoxic T cell and upregulation of inhibitory receptors on the cytotoxic T cells.[20][21] TOX promotes the exhausted T cell phenotype through epigenetic remodeling.[20][22] PD-1 is an inhibitory marker on T cells that increases when TOX is unregulated.[20][23][22] This allows for cancerous cells to evade the cytotoxic T cells through upregulated expression of PD-L1.[24]
Effector function
Markers of effector functions that are decreased when TOX is overexpressed are KLRG1, TNF, and IFN-gamma.[8] IFN-gamma and TNF-alpha production are also increased when the Tox and Tox2 genes are deleted.[9] Upregulation of effector function in cells lacking TOX is not always seen and it has been proposed that inhibitory receptor function is separated from effector CD8+ cytotoxic T cell function.[8] T-cell exhaustion does not occur when TOX is deleted from CD8+ T cells, but the cells instead adopt the KLRG1+ terminal effector state and undergo apoptosis, or programmed cell death.[9] It was therefore proposed that TOX prevents this terminal differentiation and instead promotes exhaustion so that the T-cell has a slightly more sustained response.[9]
Cancer & chronic infection
In cancer or during chronic viral infection, T-cell exhaustion occurs when cytotoxic T-cells are constantly stimulated.[8][25] TOX is upregulated in CD8+ T cells from chronic infection when compared to acute infection.[8] Patients with cancer typically have high levels of TOX in their tumor-infiltrating lymphocytes,[8] and anti-tumor immunity is heightened when Tox and Tox2 are deleted.[9] TOX and TOX2-deficient tumor-specific CAR T cells additionally have increased antitumor effector cell function as well as decreased levels of inhibitory receptors.[8]
Activation
NFAT transcription factors are essential for activating TOX in CD8+ T-cells,[8] and it has been suggested that TOX is a downstream target of NFAT.[9] The expression and function of NR4a (a target of NFAT) and TOX are strongly linked with reduced NR4a expression in Tox double knockout T cells and minimized Tox expression in NR4a triple knockout T cells.[9]
T-cell development
TOX is necessary for positive selection in developing thymocytes.[26] Knock out TOX mice shows a requirement of TOX for the CD4 T cell lineage,[26] however CD8 single positive T-cells were still able to develop.[26]
Innate lymphoid cells development
TOX is necessary for the development of innate lymphoid cells.[10][11] Innate lymphoid cells include ILC1, ILC2, ILC3 and NK cells.[26]
Notch signaling can aid in the development of all innate lymphoid cells, but in TOX-deficient cells, Notch target genes are expressed at low levels, so it is possible that TOX is required for downstream activation of these Notch target genes.[10] TOX was also found to bind Hes1, a Notch target gene, in embryonic kidney cells.[10]
Several ILC3 populations are reduced in the absence of TOX, implicating TOX’s role in their development.[10] In the small intestine, major ILC3 populations are normal in TOX-deficient cells, suggesting that gut ILC3 development may occur independently of TOX.[10] Some ILC3 populations in the gut expand in the absence of TOX.[10]
It has been proposed that NFIL3 and TOX regulate the transition of common lymphoid progenitor to early innate lymphoid progenitor.[11] In NFIL3-deficient mice, the expression of TOX is downregulated, indicating that NFIL3 is directly affecting the expression of TOX which is then acting downstream in ILC development.[11] TOX-deficient mice and NFIL3-deficient mice both lack mature ILCs and ILC progenitors.[11]
References
- GRCh38: Ensembl release 89: ENSG00000198846 - Ensembl, May 2017
- GRCm38: Ensembl release 89: ENSMUSG00000041272 - Ensembl, May 2017
- "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Miyajima N, Tanaka A, et al. (October 1998). "Prediction of the coding sequences of unidentified human genes. XI. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro". DNA Research. 5 (5): 277–286. doi:10.1093/dnares/5.5.277. PMID 9872452.
- Wilkinson B, Chen JY, Han P, Rufner KM, Goularte OD, Kaye J (March 2002). "TOX: an HMG box protein implicated in the regulation of thymocyte selection". Nature Immunology. 3 (3): 272–280. doi:10.1038/ni767. PMID 11850626. S2CID 19716719.
- "Entrez Gene: thymocyte selection-associated high mobility group box gene TOX".
- Bordon Y (August 2019). "TOX for tired T cells". Nature Reviews. Immunology. 19 (8): 476. doi:10.1038/s41577-019-0193-9. PMID 31243349.
- Ando M, Ito M, Srirat T, Kondo T, Yoshimura A (March 2020). "Memory T cell, exhaustion, and tumor immunity". Immunological Medicine. 43 (1): 1–9. doi:10.1080/25785826.2019.1698261. PMID 31822213.
- Seehus CR, Kaye J (2015). "The Role of TOX in the Development of Innate Lymphoid Cells". Mediators of Inflammation. 2015: 243868. doi:10.1155/2015/243868. PMC 4628649. PMID 26556952.
- Stokic-Trtica V, Diefenbach A, Klose CS (2020). "NK Cell Development in Times of Innate Lymphoid Cell Diversity". Frontiers in Immunology. 11: 813. doi:10.3389/fimmu.2020.00813. PMC 7360798. PMID 32733432.
- O'Flaherty E, Kaye J (April 2003). "TOX defines a conserved subfamily of HMG-box proteins". BMC Genomics. 4 (1): 13. doi:10.1186/1471-2164-4-13. PMC 155677. PMID 12697058.
- Minton K (January 2020). "TOX2 helping hand for TFH cells". Nature Reviews. Immunology. 20 (1): 4–5. doi:10.1038/s41577-019-0249-x. PMID 31745259. S2CID 208145693.
- Easton DF, Pooley KA, Dunning AM, Pharoah PD, Thompson D, Ballinger DG, et al. (June 2007). "Genome-wide association study identifies novel breast cancer susceptibility loci". Nature. 447 (7148): 1087–1093. Bibcode:2007Natur.447.1087E. doi:10.1038/nature05887. PMC 2714974. PMID 17529967.
- Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, Gudjonsson SA, et al. (July 2007). "Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer". Nature Genetics. 39 (7): 865–869. doi:10.1038/ng2064. PMID 17529974. S2CID 7346190.
- Aliahmad P, Kaye J (January 2008). "Development of all CD4 T lineages requires nuclear factor TOX". The Journal of Experimental Medicine. 205 (1): 245–256. doi:10.1084/jem.20071944. PMC 2234360. PMID 18195075.
- Alfei F, Kanev K, Hofmann M, Wu M, Ghoneim HE, Roelli P, et al. (July 2019). "TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection". Nature. 571 (7764): 265–269. doi:10.1038/s41586-019-1326-9. PMID 31207605. S2CID 190528786.
- Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, et al. (July 2019). "TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion". Nature. 571 (7764): 211–218. doi:10.1038/s41586-019-1325-x. PMC 6713202. PMID 31207603.
- Scott AC, Dündar F, Zumbo P, Chandran SS, Klebanoff CA, Shakiba M, et al. (July 2019). "TOX is a critical regulator of tumour-specific T cell differentiation". Nature. 571 (7764): 270–274. doi:10.1038/s41586-019-1324-y. PMC 7698992. PMID 31207604. S2CID 190538130.
- Bordon Y (August 2019). "TOX for tired T cells". Nature Reviews. Immunology. 19 (8): 476. doi:10.1038/s41577-019-0193-9. PMID 31243349. S2CID 195657349.
- Blank CU, Haining WN, Held W, Hogan PG, Kallies A, Lugli E, et al. (November 2019). "Defining 'T cell exhaustion'". Nature Reviews. Immunology. 19 (11): 665–674. doi:10.1038/s41577-019-0221-9. PMC 7286441. PMID 31570879.
- Cheng Y, Shao Z, Chen L, Zheng Q, Zhang Q, Ding W, et al. (January 2021). "Role, function and regulation of the thymocyte selection-associated high mobility group box protein in CD8+ T cell exhaustion". Immunology Letters. 229: 1–7. doi:10.1016/j.imlet.2020.11.004. PMID 33186634. S2CID 226948315.
- Kim K, Park S, Park SY, Kim G, Park SM, Cho JW, et al. (February 2020). "Single-cell transcriptome analysis reveals TOX as a promoting factor for T cell exhaustion and a predictor for anti-PD-1 responses in human cancer". Genome Medicine. 12 (1): 22. doi:10.1186/s13073-020-00722-9. PMC 7048139. PMID 32111241.
- van der Leun AM, Thommen DS, Schumacher TN (April 2020). "CD8+ T cell states in human cancer: insights from single-cell analysis". Nature Reviews. Cancer. 20 (4): 218–232. doi:10.1038/s41568-019-0235-4. PMC 7115982. PMID 32024970.
- Philip M, Schietinger A (July 2021). "CD8+ T cell differentiation and dysfunction in cancer". Nature Reviews. Immunology. 22 (4): 209–223. doi:10.1038/s41577-021-00574-3. PMC 9792152. PMID 34253904. S2CID 235808117.
- Aliahmad P, Seksenyan A, Kaye J (April 2012). "The many roles of TOX in the immune system". Current Opinion in Immunology. 24 (2): 173–177. doi:10.1016/j.coi.2011.12.001. PMC 3319641. PMID 22209117.
Further reading
- Nakajima D, Okazaki N, Yamakawa H, Kikuno R, Ohara O, Nagase T (June 2002). "Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones". DNA Research. 9 (3): 99–106. doi:10.1093/dnares/9.3.99. PMID 12168954.
- Sebastiani P, Wang L, Nolan VG, Melista E, Ma Q, Baldwin CT, Steinberg MH (March 2008). "Fetal hemoglobin in sickle cell anemia: Bayesian modeling of genetic associations". American Journal of Hematology. 83 (3): 189–195. doi:10.1002/ajh.21048. PMID 17918249. S2CID 24667609.
- Aliahmad P, O'Flaherty E, Han P, Goularte OD, Wilkinson B, Satake M, et al. (April 2004). "TOX provides a link between calcineurin activation and CD8 lineage commitment". The Journal of Experimental Medicine. 199 (8): 1089–1099. doi:10.1084/jem.20040051. PMC 2211890. PMID 15078895.