Microbacterium virus MuffinTheCat
Microbacterium virus MuffinTheCat is a species of bacteriophage in the family Tectiviridae. It was collected and identified by Darcy Reimer on 1 October 2019.[1][2] It is part of the Microbacterium testaceum NRRL B-24232 viral strand and the GE viral cluster. Microbacterium of the Microbacterium testaceum species serve as natural hosts. Microbacterium virus MuffinTheCat is morphologically almost indistinguishable from its sibling species in the Tectiviridae family,[2] but it along with its sibling species in the GE cluster are different enough from other Tectiviridae members that the GE cluster may soon be identified as a new genus.[3] Microbacterium virus MuffinTheCat is identified from other GE cluster members by its genome differences.[2][4]
| Microbacterium virus MuffinTheCat | |
|---|---|
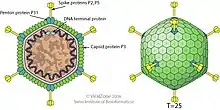 | |
| Internal and external diagram of, Bacillus virus Bam35, in the Tectviridae Family | |
| Scientific classification | |
| (unranked): | Virus |
| Realm: | Varidnaviria |
| Kingdom: | Bamfordvirae |
| Phylum: | Preplasmiviricota |
| Class: | Tectiliviricetes |
| Order: | Kalamavirales |
| Family: | Tectiviridae |
| Species: | M. v. MuffinTheCat |
| Binomial name | |
| Microbacterium virus MuffinTheCat Darcy Reimer - 2019 | |
| Synonyms | |
|
- Microbacterium virus Jeffery (Former) - Microbacterium phage MuffinTheCat (Synonym) | |
Morphology
Family specific morphology
Microbacterium virus MuffinTheCat shows morphological similarity to other Tectiviridae. They are non-enveloped, icosahedral and have T=25 symmetry, (how the proteins on their surface are arranged, see[5]). They have a virion size of approximately 66 nm and apical spikes of approximately 20 nm. Tectiviridae have a genome of approximately 15kb.[6]
Specific genome identification
Microbacterium virus MuffinTheCat however, has a genome size of exactly 15494bp (15.494kb) coding for 32 genes and 30 ORFs. This dsDNA genome is flanked by inverted repeats and DNA replication is protein primed.[2] The GC-content of Microbacterium virus MuffinTheCat's genome is 55.1%.[2] GC-content relates to how strong an entitiy's DNA is, as a higher GC-content means more hydrogen bonds and a structurally stronger genome.[7] Comparatively, most of its sibling species such as Microbacterium virus Badulia have similar genome sizes, but slightly higher GC-content, with Microbacterium virus Badulia having 54.6% GC-content and Microbacterium virus Franklin22 (EB cluster) having 66.1%.[8]
Sequencing
Genome sequencing of Microbacterium virus MuffinTheCat was conducted by the Pittsburgh Bacteriophage Institute on Dec 12, 2019. It was done using the Illumina shotgun sequencing method. With a total shotgun coverage (amount of sequencing material) of 824. It was during this prosses that information such as genome size and GC-content was uncovered.[2]
Discovery and habitat
Discovery

Microbacterium virus MuffinTheCat was discovered by Darcy Reimer in 2019 in Nyack, New York, United States, from an enriched soil sample from a moist area of a mostly dried-up riverbed. It was discovered via the Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science Program at Nyack College.[2]
Habitat
Its host species Microbacterium testaceum (Strain, NRRL B-24232) infects plants such as potatoes or rice,[9] this interspecific relationship is relatively commensal. Therefore it is presumed that Microbacterium virus MuffinTheCat infects strands or even reproductively isolated subspecies of Microbacterium testaceum that harbour freshwater plant life or deep-rooted riverside plants.[10] However, this is relatively speculative as only one known sample has been taken of this bacteriophage species.[2]
Naming
Microbacterium virus MuffinTheCat was originally named Microbacterium virus Jeffery (Former name), an unknown member of the discovery project states "Jeffrey is a good name for a phage, picked randomly."[2] However, this name was changed, likely to be more meaningful. It is evident from scientific classifications such as Streptomyces phage Forthebois which was a name emerging from the same identification project. That the college students that were a part of this program naturally got to name the species they discovered.[1] Muffin the cat is likely the identifier Darcy Reimer's cat (Felidae) or a domesticated cat that is close to them. This claim is backed up by the naming of Mycobacterium phage DaisytheDog also identified by Nyack college with the naming notes stating, "My friend's dog who looks a lot like Air Bud is the reason for the name."[11] This shows the naming of Microbacterium virus MuffinTheCat was likely not random and was named after a cat called Muffin. Disregarding the species Pellonula leonensis which has a common name; Muffin sprat, Microbacterium virus MuffinTheCat is one of 2 species with "Muffin" in its scientific classification. The other being a mollusk in the Buccinidae family, Muffinbuccinum catherinae.[12]
Taxonomy
Divergence
Microbacterium virus MuffinTheCat is in the family Tectiviridae, and the Kingdom Bamfordvirae. The average nucleotide identity (ANI), that was measured on each of the GE cluster species shows that members of the GE cluster are <60% similar to other members of the Tectiviridae family. Such a low ANI score indicates clear taxonomic divergence from the other Tectiviridae genera. This coupled with the fact that the members of the GE cluster are distinct from other Tectiviridae genera discovered thus far, shows that the GE cluster should be a new genus.[3] Such clear genetic difference indicates that Microbacterium virus MuffinTheCat and its sibling species taxonomically diverged from other Tectiviridae a long time ago. By using a crude (genetic difference over mutation rate calculation) we can achieve a rough taxonomic timeline. Bacteriophages have a life cycle of approximately 30 minutes.[13] Most viruses have a mutation rate of approximately one mutation every several hundred to many thousand generations, a value of 500 is a safe pick.[14] Here the mutation rate can be identified at roughly 1 mutation every 11 days. This is backed up by claims from virologist Louis Mansky, who states the mutation rate of viruses like COVID-19 are about once every 11 days, mutation rates between virus species do not differ largely.[15] Considering a virus to compare to such as Bacillus virus AP50 from the Tectiviridae family, which has a genome of 14kb.[16] With a similarity of 60% as stated above you get a genetic difference of 5600 mutations, 5600 * 11 days = ~169 years. This is a very long time for taxonomic divergence in the virus realm. As viruses reproduce so effectively with little regard to genetic conservation. Although these calculations are very crude and full of assumptions, they show the point that the GE cluster containing Microbacterium virus MuffinTheCat are a clear genus that has diverged from other cousin species a long time ago.
Taxonomic relatives
Parent Genus:
- Unclassified
Parent Family:
Kingdom:
Sibling Strand Species:
GE cluster:
- Microbacterium phage Badulia
- Microbacterium phage DesireeRose
- Microbacterium phage LuzDeMundo
EB cluster:
- Microbacterium phage Franklin22
Undefined cluster: (Not sequenced)
- Microbacterium phage Bee17
- Microbacterium phage CharleeAnn
- Microbacterium phage Gargantuan
- Microbacterium phage Plimp
- Microbacterium phage SCoupsA
- Microbacterium phage Sticker
Sibling and Cousin Species and Their Genera (Under Tectiviridae):
- Alphatectivirus
- Pseudomonas virus PR4
- Pseudomonas virus PRD1
- Betatectivirus
- Bacillus virus AP50
- Bacillus virus Bam35
- Bacillus virus GIL16
- Bacillus virus Wip1
- Deltatectivirus
- Streptomyces virus Forthebois
- Streptomyces virus WheeHeim
- Epsilontectivirus
- Rhodococcus virus Toil
- Gammatectivirus
- Gluconobacter virus GC1
- Undefined
- Thermus virus phiKo
References
- "Microbacterium phage MuffinTheCat, complete genome". Ncbi.nlm.nih.gov. 2021-12-02.
- "The Actinobacteriophage Database | Phage MuffinTheCat". Phagesdb.org. Retrieved 2022-07-06.
- "SEA-PHAGES | Abstracts". Seaphages.org. Retrieved 2022-07-06.
- Tse, Vincent Y.; Liu, Haoyuan; Kapinos, Andrew; Torres, Canela; Cayabyab, Breanna Camille S.; Fett, Sarah N.; Nakashima, Lucy G.; Rahman, Mujtahid; Vargas, Aida S.; Reddi, Krisanavane; Parker, Jordan Moberg; Freise, Amanda C. (2022). "Exploring Genomic Conservation in Actinobacteriophages with Small Genomes" (PDF). Biorxiv.org. doi:10.1101/2022.02.05.479200. S2CID 246653340. Retrieved 16 July 2022.
- "T=25 icosahedral capsid protein ~ ViralZone". Viralzone.expasy.org. Retrieved 2022-07-06.
- "Tectiviridae ~ ViralZone". Viralzone.expasy.org. Retrieved 2022-07-06.
- Chauhan, Dr Tushar (2021-08-25). "What is the importance of GC Content?". Genetic Education. Retrieved 2022-07-06.
- "The Actinobacteriophage Database| Microbacterium testaceum NRRL B-24232". Phagesdb.org. Retrieved 2022-07-06.
- "Microbacterium testaceum NRRL B-24232 (Type Strain)". Brrl.ncaur.usda.gov. Retrieved 16 July 2022.
- Morohoshi, Tomohiro; Wang, Wen-Zhao; Someya, Nobutaka; Ikeda, Tsukasa (April 2011). "Genome Sequence of Microbacterium testaceum StLB037, an N-Acylhomoserine Lactone-Degrading Bacterium Isolated from Potato Leaves▿". Journal of Bacteriology. 193 (8): 2072–2073. doi:10.1128/JB.00180-11. ISSN 0021-9193. PMC 3133038. PMID 21357489.
- "The Actinobacteriophage Database | Phage DaisytheDog". Phagesdb.org. Retrieved 2022-07-06.
- "Muffinbuccinum catherinae Harasewych & Kantor, 2004 | COL". Catalogueoflife.org. Retrieved 2022-07-06.
- "Bacteriophage". Textbookofbacteriology.net. Retrieved 2022-07-06.
- Fleischmann, W. Robert (1996), Baron, Samuel (ed.), "Viral Genetics", Medical Microbiology (4th ed.), Galveston (TX): University of Texas Medical Branch at Galveston, ISBN 978-0-9631172-1-2, PMID 21413337, retrieved 2022-07-06
- "The coronavirus is mutating—but what determines how quickly?". Science. 2021-02-05. Retrieved 2022-07-07.
- Sozhamannan, Shanmuga; McKinstry, Michael; Lentz, Shannon M.; Jalasvuori, Matti; McAfee, Farrell; Smith, Angela; Dabbs, Jason; Ackermann, Hans-W.; Bamford, Jaana K. H.; Mateczun, Alfred; Read, Timothy D. (November 2008). "Molecular Characterization of a Variant of Bacillus anthracis-Specific Phage AP50 with Improved Bacteriolytic Activity". Applied and Environmental Microbiology. 74 (21): 6792–6796. Bibcode:2008ApEnM..74.6792S. doi:10.1128/AEM.01124-08. ISSN 0099-2240. PMC 2576697. PMID 18791014.
External links
- Taxonomy Browser: Taxonomy browser (Microbacterium phage MuffinTheCat)
- Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science Program: SEA-PHAGES | Home (Copywrite holder of Microbacterium virus MuffinTheCat images)