Cystovirus
Cystovirus is a genus of double-stranded RNA viruses which infects bacteria. It is the only genus in the family Cystoviridae. The name of the group cysto derives from Greek kystis which means bladder or sack. There are seven species in this genus.[1][2][3]
| Cystovirus | |
|---|---|
 | |
 | |
| Pseudomonas phage phi6 particle, reconstruction, and genome | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Duplornaviricota |
| Class: | Vidaverviricetes |
| Order: | Mindivirales |
| Family: | Cystoviridae |
| Genus: | Cystovirus |
| Species | |
Discovery
Pseudomonas virus phi6 was the first virus in this family to be discovered and was initially characterized in 1973 by Anne Vidaver at the University of Nebraska. She found that when she cultured the bacterial strain Pseudomonas phaseolicola HB1OY with halo blight infected bean straw, cytopathic effects were detected in cultured lawns, indicating that there was a lytic microbe or bacteriophage present.[4]
In 1999, phi7–14 were identified by the laboratory of Leonard Mindich at the Public Health Research Institute associated with New York University. They did this by culturing various leaves in Lysogeny Broth and then plating the broth on lawns of Pseudomonas syringae pv phaseolicola. They were able to identify viral plaques from this and then subsequently sequence their genomes.[5]
Microbiology
Structure
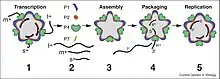
Cystovirus particles are enveloped, with icosahedral and spherical geometries, and T=13, T=2 symmetry. The virion diameter is around 85 nm. Cystoviruses are distinguished by their outer layer protein and lipid envelope. No other bacteriophage has any lipid in its outer coat, though the Tectiviridae and the Corticoviridae have lipids within their capsids.[1][2]
Genome
Cystoviruses have a tripartite double-stranded RNA genome which is approximately 14 kbp in total length. The genome is linear and segmented, and labeled as large (L) 6.4 kbp, medium (M) 4 kbp, and small (S) 2.9 kb in length. The genome codes for twelve proteins.[1][2]
Life cycle
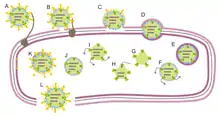
Cytoviruses enter the bacteria by adsorption on its pilus and then membrane fusion. Viral replication is cytoplasmic. Replication follows the double-stranded RNA virus replication model. Double-stranded RNA virus transcription is the method of transcription. The progeny viruses are released from the cell by lysis.[1][2]
Most identified cystoviruses infect Pseudomonas species, but this is likely biased due to the method of screening and enrichment.[6] There are many proposed members of this family. Pseudomonas viruses φ7, φ8, φ9, φ10, φ11, φ12, and φ13 have been identified and named,[5] but other cystovirus-like viruses have also been isolated.[6] These seven putative relatives are classified as either close (φ7, φ9, φ10, φ11) or distant (φ8, φ12, φ13) relatives to φ6,[5] with the distant relatives thought to infect via the LPS rather than the pili.[7]
However, cystoviruses do not only infect Pseudomonas. But also bacteria of the genera Streptomyces,[8] Microvirgula,[9] Acinetobacter,[10] Lactococcus, Pectobacterium,[11] and possibly other bacterial genera.
Taxonomy
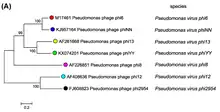
Members of the Cystoviridae appear to be most closely related to the Reoviridae,[12] but also share homology with the Totiviridae. In particular, the structural genes of cystoviruses are highly-similar to those used by a number of dsRNA viruses that infect eukaryotes.[13] The genus Cystovirus has seven species:[1]
- Pseudomonas virus phi6
- Pseudomonas virus phi8
- Pseudomonas virus phi12
- Pseudomonas virus phi13
- Pseudomonas virus phi2954
- Pseudomonas virus phiNN
- Pseudomonas virus phiYY
Other unassigned phages:
References
- "ICTV Report Cystoviridae".
- "Viral Zone". ExPASy. Retrieved 15 June 2015.
- "NCBI Taxonomy Browser: Cystoviridae". NCBI. Retrieved 19 June 2016.
- Vidaver AK, Koski RK, Van Etten JL (May 1973). "Bacteriophage Φ6 a Lipid-Containing Virus of Pseudomonas phaseolicola". Journal of Virology. 11 (15): 799–805. doi:10.1128/jvi.11.5.799-805.1973. PMC 355178. PMID 16789137.
- Mindich L, Qiao X, Qiao J, Onodera S, Romantschuk M, Hoogstraten D (August 1999). "Isolation of additional bacteriophages with genomes of segmented double-stranded RNA". J. Bacteriol. 181 (15): 4505–8. doi:10.1128/JB.181.15.4505-4508.1999. PMC 103579. PMID 10419946.
- Silander OK, Weinreich DM, Wright KM, et al. (December 2005). "Widespread genetic exchange among terrestrial bacteriophages". Proc. Natl. Acad. Sci. U.S.A. 102 (52): 19009–14. Bibcode:2005PNAS..10219009S. doi:10.1073/pnas.0503074102. PMC 1323146. PMID 16365305.
- Gottlieb P, Potgieter C, Wei H, and Toporovsky I (2002). "Characterization of φ12, a Bacteriophage Related to φ6: Nucleotide Sequence of the Large Double-Stranded RNA". Virology. 295 (2): 266–271. doi:10.1006/viro.2002.1436. PMID 12033785.
- Siddharth R. Krishnamurthy, Andrew B. Janowski,Guoyan Zhao, Dan Barouch, David Wang (2016). Hyperexpansion of RNA Bacteriophage Diversity. PlosOne.
- Xiaoyao Cai, Fengjuan Tian, Li Teng, Hongmei Liu, Yigang Tong Tong, Shuai Le, Tingting Zhang (2021). Cultivation of a Lytic Double-Stranded RNA Bacteriophage Infecting Microvirgula aerodenitrificans Reveals a Mutualistic Parasitic Lifestyle. American Society for Microbiology.
- Clay S. Crippen, Bibi Zhou, and Christine M. Szymanski (2021). RNA and Sugars, Unique Properties of Bacteriophages Infecting Multidrug Resistant Acinetobacter radioresistens Strain LH6. NCBI.
- Cystoviridae. NCBI Taxonomy.
- Butcher SJ, Dokland T, Ojala PM, Bamford DH, Fuller SD (July 1997). "Intermediates in the assembly pathway of the double-stranded RNA virus phi6". EMBO J. 16 (14): 4477–87. doi:10.1093/emboj/16.14.4477. PMC 1170074. PMID 9250692.
- Koonin, Eugene V.; Dolja, Valerian V.; Krupovic, Mart (2015). "Origins and evolution of viruses of eukaryotes: The ultimate modularity". Virology. 479–480: 2–25. doi:10.1016/j.virol.2015.02.039. PMC 5898234. PMID 25771806.