Phenylethanolamine N-methyltransferase
| phenylethanolamine N-methyltransferase | |||||||
|---|---|---|---|---|---|---|---|
 Phenylethanolamine N-methyltransferase monomer, Human | |||||||
| Identifiers | |||||||
| Symbol | PNMT | ||||||
| Alt. symbols | PENT | ||||||
| NCBI gene | 5409 | ||||||
| HGNC | 9160 | ||||||
| OMIM | 171190 | ||||||
| RefSeq | NM_002686 | ||||||
| UniProt | P11086 | ||||||
| Other data | |||||||
| EC number | 2.1.1.28 | ||||||
| Locus | Chr. 17 q21-q22 | ||||||
| |||||||
Phenylethanolamine N-methyltransferase (PNMT) is an enzyme found primarily in the adrenal medulla that converts norepinephrine (noradrenaline) to epinephrine (adrenaline).[1] It is also expressed in small groups of neurons in the human brain[2] and in selected populations of cardiomyocytes.[3]
Structure
PNMT is a protein whose encoding gene is found on chromosome 17 in humans. It consists of 4 exons and is a 30kDa protein. It shares many properties found among the other methyltransferases. It is closest in sequence to glycine-N-methyl transferase (GNMT). It also shares many structural properties like the shape of the folding lip with catechol-O-methyl transferase (COMT), though it shares less sequence identity. Several features of the structure like this folding lip suggest that PNMT is a recent adaptation to the catecholamine synthesizing enzyme family, evolving later than COMT, but before other methyltransferases like GNMT.[5]
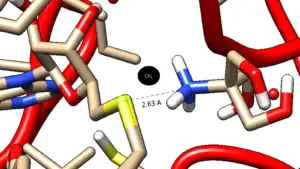
S-adenosyl-L-methionine (SAM) is a required cofactor.[6] The active site binding region for the cofactor SAM contains a rich number of pi bonds from phenylalanine and tyrosine residues in the active site help to keep it in its binding pocket through pi stacking. Among all known PNMT variants in nature there are 7 crucial aromatic residues conserved in the active site.[5]
The residue Glutamine 185 is necessary in binding the catecholamine substrate. The replacement of this residue another reduces the catalytic efficiency of PNMT by tenfold up to three hundredfold.[7]
In the absence of an inhibitor or ligand, a phosphate group is bound to the active site to stabilize this region.[8]
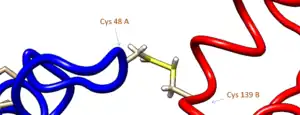
Human PNMT forms dimers in solution. When PNMT crystals are grown in non-reducing solutions, two disulfide bonds form between cysteines 48 and 139 on opposite chains. This dimerization has no effect on the catalytic activity of the enzyme.[9]
Mechanism
PNMT catalyzes the transfer of a methyl group from SAM to norepinephrine, converting it into epinephrine. It works by bringing the cofactor SAM and substrate together in close proximity, so that the reactive methyl group can be attacked by the primary amine of the norepinephrine molecule or another catecholamine substrate. The methyl group of SAM is very reactive, so the structure and placement of both norepinephrine and SAM is crucial for correct methylation pattern on the product.
While PNMT methylates norepinephrine into the active compound epinephrine, norepinephrine can also be methylated by catechol-O-methyl transferase (COMT), another methyltransferase which adds a methyl group in a different location, in turn producing the inactive compound metanephrine. Methyltransferases are very common in the catecholamine synthesis and deactivation pathways.[10]
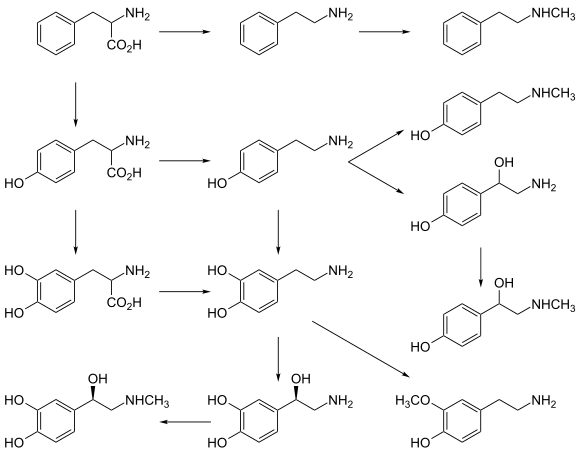
PNMT is also involved in the biosynthesis of N-methylated trace amines: it metabolizes phenethylamine into N-methylphenethylamine (a positional isomer of amphetamine), p-octopamine into synephrine, and p-tyramine into N-methyltyramine.[11][12]
Regulation
Elevated PNMT expression is one of the ways that the stress response positively feeds back on itself. An increase in stress hormones or nerve impulses due to stress can cause PNMT to convert more norepinephrine into epinephrine. This increases the potency of the catecholamine response system, increasing the sympathetic output and making the stress response more profound.[14]
PNMT is known to be regulated by glucocorticoids made in the adrenal gland. One way that it can regulate PNMT expression is by corticosterone's positive influence on the maintenance of PNMT mRNA.[15] Glucocorticoids have also been shown to increase the biological half life of the enzyme in vitro.[16] In animals who have had their pituitary gland removed, the addition of glucocorticoids significantly lengthens the half life of PNMT enzymes.[16]
Elevated PNMT levels can also be triggered by splanchnic nerve impulses. Nerve impulses increase the synthesis of PNMT mRNA by affecting certain promoter sequences.[16]
Stress immobilization for a few hours has also been shown to increase PNMT activity in rats. This treatment takes about one week to manifest a difference in PNMT levels.[17]
SAM not only acts as a cofactor for PNMT, but also helps to stabilize the enzyme and increase the half life by making it more resistant to being cut by trypsin protease.[16]
Localization
Epinephrine synthesis and therefore PNMT location has been largely found to be contained in the adrenal medulla or adrenal gland of most species. PNMT has been localized in most adult mammals to the cytoplasm of these medullary cells.[1]
Newer studies are also showing PNMT mRNA and protein to be expressed in other regions of the body as well. Certain neural tracts, the retina,[18] and in both atria and ventricles in the hearts are now being elucidated as sites of PNMT expression.[19] Epinephrine is produced in small groups of neurons in the human brain which express PNMT;[2] these neurons project from a nucleus that is adjacent (ventrolateral) to the area postrema and from a nucleus in the dorsal region of the solitary tract.[2]
Disease
PNMT's normal function and defects are associated with multiple diseases and disorders.
Vitiligo
Decreased levels of PNMT activity measured by epinephrine and norepinephrine is seen in the skin of patients with vitiligo in the keratinocytes, which normally have higher PNMT activity.[20]
Ethanol intoxication
Two potent PNMT inhibitors (LY134046 and LY78335) were long lasting antagonists of both ethanol intoxication and sedation. This suggests a central role that PNMT and epinephrine play in the synthesis of ethanol and pentobarbital induced sedation and intoxication.[21]
Alzheimer's disease
Alzheimer's disease has also been associated with reduced human PNMT activity in the regions of the brain most associated with degeneration in the disease. There have also been significant associations with PNMT polymorphisms and early onset Alzheimer's disease.[22]
Inhibition
Classic PNMT inhibitors include benzimidazoles, quinolones, and purines.[8] Inhibition can also be produced by the addition of S-deoxyadenosyl L-homocysteine, a replacement for the cofactor SAM, which resembles it, but is missing the methyl group, so no methyl transfer is possible.[23]
References
- 1 2 Goldstein M, Fuxe K, Hökfelt T (June 1972). "Characterization and tissue localization of catecholamine synthesizing enzymes". Pharmacological Reviews. 24 (2): 293–309. PMID 4564603.
- 1 2 3 Kitahama K, Pearson J, Denoroy L, Kopp N, Ulrich J, Maeda T, Jouvet M (February 1985). "Adrenergic neurons in human brain demonstrated by immunohistochemistry with antibodies to phenylethanolamine-N-methyltransferase (PNMT): discovery of a new group in the nucleus tractus solitarius". Neuroscience Letters. 53 (3): 303–8. doi:10.1016/0304-3940(85)90555-5. PMID 3885079. S2CID 2578817.
- ↑ Wang Y, Lin WK, Crawford W, Ni H, Bolton EL, Khan H, et al. (January 2017). "+ Cells in Murine Heart". Scientific Reports. 7 (1): 40687. doi:10.1038/srep40687. PMC 5234027. PMID 28084430.
- 1 2 PDB: 4MQ4; Bart AG, Scott EE. Crystal Structure of hPNMT in Complex with bisubstrate inhibitor N-(3-((((2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)thio)propyl)-1,2,3,4-tetrahydroisoquinoline-3-carboxamide (Report). RCSB Protein Data Bank.
- 1 2 Martin JL, Begun J, McLeish MJ, Caine JM, Grunewald GL (October 2001). "Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT". Structure. 9 (10): 977–85. doi:10.1016/s0969-2126(01)00662-1. PMID 11591352.
- ↑ Wong DL, Lesage A, Siddall B, Funder JW (November 1992). "Glucocorticoid regulation of phenylethanolamine N-methyltransferase in vivo". FASEB Journal. 6 (14): 3310–5. doi:10.1096/fasebj.6.14.1426768. PMID 1426768. S2CID 23761885.
- ↑ Drinkwater N, Gee CL, Puri M, Criscione KR, McLeish MJ, Grunewald GL, Martin JL (August 2009). "Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity". The Biochemical Journal. 422 (3): 463–71. doi:10.1042/bj20090702. hdl:1808/26489. PMC 5940352. PMID 19570037.
- 1 2 Drinkwater N, Vu H, Lovell KM, Criscione KR, Collins BM, Prisinzano TE, et al. (October 2010). "Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors". The Biochemical Journal. 431 (1): 51–61. doi:10.1042/bj20100651. PMID 20642456.
- ↑ Gee CL, Nourse A, Hsin AY, Wu Q, Tyndall JD, Grunewald GL, et al. (June 2005). "Disulfide-linked dimers of human adrenaline synthesizing enzyme PNMT are catalytically active". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 1750 (1): 82–92. doi:10.1016/j.bbapap.2005.03.006. PMID 15893506.
- ↑ Brandt. "The Adrenal Medulla" (PDF).
- 1 2 Broadley KJ (March 2010). "The vascular effects of trace amines and amphetamines". Pharmacology & Therapeutics. 125 (3): 363–375. doi:10.1016/j.pharmthera.2009.11.005. PMID 19948186.
- 1 2 Lindemann L, Hoener MC (May 2005). "A renaissance in trace amines inspired by a novel GPCR family". Trends in Pharmacological Sciences. 26 (5): 274–281. doi:10.1016/j.tips.2005.03.007. PMID 15860375.
- ↑ Wang X, Li J, Dong G, Yue J (February 2014). "The endogenous substrates of brain CYP2D". European Journal of Pharmacology. 724: 211–218. doi:10.1016/j.ejphar.2013.12.025. PMID 24374199.
- ↑ Wurtman RJ (June 2002). "Stress and the adrenocortical control of epinephrine synthesis". Metabolism. 51 (6 Suppl 1): 11–4. doi:10.1053/meta.2002.33185. PMID 12040535.
- ↑ Jiang W, Uht R, Bohn MC (1989). "Regulation of phenylethanolamine N-methyltransferase (PNMT) mRNA in the rat adrenal medulla by corticosterone". International Journal of Developmental Neuroscience. 7 (5): 513–20. doi:10.1016/0736-5748(89)90010-5. PMID 2816488. S2CID 24803398.
- 1 2 3 4 Ciaranello RD (1978). "Regulation of phenylethanolamine N-methyltransferase". Biochemical Pharmacology. 27 (15): 1895–7. doi:10.1016/0006-2952(78)90002-3. PMID 708473.
- ↑ Cahill AL, Eertmoed AL, Mangoura D, Perlman RL (September 1996). "Differential regulation of phenylethanolamine N-methyltransferase expression in two distinct subpopulations of bovine chromaffin cells". Journal of Neurochemistry. 67 (3): 1217–24. doi:10.1046/j.1471-4159.1996.67031217.x. PMID 8752129.
- ↑ Park DH, Teitelman G, Evinger MJ, Woo JI, Ruggiero DA, Albert VR, et al. (April 1986). "Phenylethanolamine N-methyltransferase-containing neurons in rat retina: immunohistochemistry, immunochemistry, and molecular biology". The Journal of Neuroscience. 6 (4): 1108–13. doi:10.1523/JNEUROSCI.06-04-01108.1986. PMC 6568425. PMID 2871139.
- ↑ Krizanová O, Micutková L, Jeloková J, Filipenko M, Sabban E, Kvetnanský R (September 2001). "Existence of cardiac PNMT mRNA in adult rats: elevation by stress in a glucocorticoid-dependent manner". American Journal of Physiology. Heart and Circulatory Physiology. 281 (3): H1372-9. doi:10.1152/ajpheart.2001.281.3.H1372. PMID 11514309.
- ↑ Schallreuter KU, Wood JM, Pittelkow MR, Buttner G, Swanson N, Korner C, Ehrke C (1996). "Increased monoamine oxidase A activity in the epidermis of patients with vitiligo". Archives of Dermatological Research. 288 (1): 14–8. doi:10.1007/bf02505037. PMID 8750929. S2CID 31646987.
- ↑ Mefford IN, Lister RG, Ota M, Linnoila M (February 1990). "Antagonism of ethanol intoxication in rats by inhibitors of phenylethanolamine N-methyltransferase". Alcoholism, Clinical and Experimental Research. 14 (1): 53–7. doi:10.1111/j.1530-0277.1990.tb00446.x. PMID 2178473.
- ↑ Mann MB, Wu S, Rostamkhani M, Tourtellotte W, MacMurray J, Comings DE (May 2001). "Phenylethanolamine N-methyltransferase (PNMT) gene and early-onset Alzheimer disease". American Journal of Medical Genetics. 105 (4): 312–6. doi:10.1002/ajmg.1363. PMID 11378842.
- ↑ Borchardt RT, Wu YS (March 1975). "Potential inhibitors of S-adenosylmethionine-dependent methyltransferases. 3. Modifications of the sugar portion of S-adenosylhomocysteine". Journal of Medicinal Chemistry. 18 (3): 300–4. doi:10.1021/jm00237a018. PMID 1133821.
External links
- Phenylethanolamine+N-Methyltransferase at the US National Library of Medicine Medical Subject Headings (MeSH)