Proteus mirabilis
| Proteus mirabilis | |
|---|---|
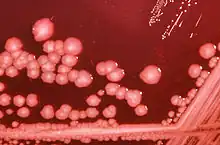 | |
| P. mirabilis on an XLD agar plate. | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Pseudomonadota |
| Class: | Gammaproteobacteria |
| Order: | Enterobacterales |
| Family: | Enterobacteriaceae |
| Genus: | Proteus |
| Species: | P. mirabilis |
| Binomial name | |
| Proteus mirabilis Hauser, 1885 | |
Proteus mirabilis is a Gram-negative, facultatively anaerobic, rod-shaped bacterium. It shows swarming motility and urease activity. P. mirabilis causes 90% of all Proteus infections in humans. It is widely distributed in soil and water.[1] Proteus mirabilis can migrate across the surface of solid media or devices using a type of cooperative group motility called swarming. Proteus mirabilis is most frequently associated with infections of the urinary tract, especially in complicated or catheter-associated urinary tract infections.[2]
Characteristics
P. mirabilis can use urea.[3] It can produce hydrogen sulfide gas, and forms clear films on growth media. It is motile, possessing peritrichous flagella, and is known for its swarming ability. It is commonly found in the human digestive system. P. mirabilis is not pathogenic in guinea pigs or chickens . This species' ability to inhibit growth of unrelated strains had been a topic for scientific curiosity, which then resulted in the discovery a macroscopically visible line of reduced bacterial growth where two swarming strains intersect. This line is named the Dienes line after its discoverer Louis Dienes.[4]
The micro-organism tests:
- Indole-negative and nitrate reductase-positive (no gas bubbles produced)
- Methyl red-positive and Voges-Proskauer negative (Can be both MR- and V-P-positive)
- Catalase positive and cytochrome oxidase-negative
- Phenylalanine deaminase-positive
- Tryptophan test-negative
- Urea test- positive
- Casein test-negative
- Starch test- negative
- Hydrogen sulfide test-positive
- Citrate agar test-positive
- Ornithine decarboxylase-positive
- Lysine decarboxylase-positive
- No fermentation of arabinose, sorbitol and dulcitol
.png.webp) Proteus mirabilis (O18 pDsRed2 biofilm) stained with live/dead BacLight stain
Proteus mirabilis (O18 pDsRed2 biofilm) stained with live/dead BacLight stain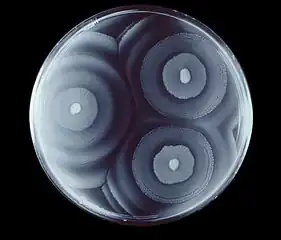 Proteus mirabilis in trypticase soy agar growth media
Proteus mirabilis in trypticase soy agar growth media Proteus mirabilis appears as Gram-negative rods after Gram staining under bright-field microscopy with 1000 times magnification.
Proteus mirabilis appears as Gram-negative rods after Gram staining under bright-field microscopy with 1000 times magnification.
Swarming motility
Swarming is a specialized form of motility that groups of multicellular, flagellated bacteria can undergo to expand their populations to new locations.[5] The swarming capability of Proteus mirabilis is important because it is implicated in the pathogenesis of the bacteria and the swarming capability is associated with the bacteria's ability to express virulence factors[6] Proteus mirabilis has a very characteristic bulls-eye appearance on an agar plate due to the regular periodic cycling between the vegetative and swarming state of the cells.[7]
In liquid culture, Proteus mirabilis exists as a vegetative cell that is approximately 2µm long and has four to ten peritrichous flagella. In the vegetative cell the flagella are used to propel the bacterium forward. Swarming cells are only formed when the bacteria are grown on solid surfaces so the ability to detect these solid surfaces is a required feature. It has been proposed that Proteus mirabilis senses a solid surface by the inhibition of its flagellum rotation, and it is this lack of freely rotating flagella that let the bacteria know it is on a solid surface. When Proteus mirabilis encounters a solid surface, and other necessary conditions have been met, the cell will undergo the differentiation process into a swarmer cell. This differentiation process includes the elongation of the cell 20 to 50 times longer than the vegetative cell, multinucleation, and more than a 50-fold greater surface density of flagella.[5]
The swarming process continues as periodic cycles of cell differentiation, population migration, and consolidation as the bacteria undergo these changes in response to environmental stimulants. The repetition of this cycle is what gives Proteus mirabilis its distinctive bulls-eye pattern when growing on solid media. This pattern can be used to distinguish Proteus mirabilis from other species of swarming bacteria. Each ring is formed when the bacteria is in the consolidation stage and the bacteria is increasing in population.[6]
Disease
This rod-shaped bacterium has the ability to produce high levels of urease, which hydrolyzes urea to ammonia (NH3), so makes the urine more alkaline. If left untreated, the increased alkalinity can lead to the formation of crystals of struvite, calcium carbonate, and/or apatite, which can result in kidney stones. The bacterium can be found throughout the stones, and these bacteria lurking in the kidney stones can reinitiate infection after antibiotic treatment. Once the stones develop, over time they may grow large enough to cause obstruction and kidney failure. Proteus species can also cause wound infections, sepsis, and pneumonia, mostly in hospitalized patients.[8]
Diagnosis
An alkaline urine sample is a possible sign of P. mirabilis. It can be diagnosed in the lab due to characteristic swarming motility, and inability to metabolize lactose (on a MacConkey agar plate, for example). Also P. mirabilis produces a very distinct fishy odor.
Treatment
P. mirabilis is generally susceptible to most antibiotics apart from tetracycline and nitrofurantoin,[9] but 10–20% of P. mirabilis strains are also resistant to first-generation cephalosporins and ampicillin.[10]
References
- ↑ "Proteus mirabilis". BioMedHTC. Archived from the original on 26 September 2009.
Bacteria of the species Proteus mirabilis are widely distributed in soil and water in the natural environment. In humans, Proteus is found as part of the normal flora of the gut....from
- ↑ Chen CY, Chen YH, Lu PL, Lin WR, Chen TC, Lin CY (June 2012). "Proteus mirabilis urinary tract infection and bacteremia: risk factors, clinical presentation, and outcomes". Journal of Microbiology, Immunology, and Infection = Wei Mian Yu Gan Ran Za Zhi. 45 (3): 228–236. doi:10.1016/j.jmii.2011.11.007. PMID 22572004.
- ↑ Schaffer JN, Pearson MM (October 2015). Mulvey MA, Stapleton AE, Klumpp DJ (eds.). "Proteus mirabilis and Urinary Tract Infections". Microbiology Spectrum. 3 (5): 3.5.10. doi:10.1128/microbiolspec.UTI-0017-2013. PMC 4638163. PMID 26542036.
- ↑ Budding AE, Ingham CJ, Bitter W, Vandenbroucke-Grauls CM, Schneeberger PM (June 2009). "The Dienes phenomenon: competition and territoriality in Swarming Proteus mirabilis". Journal of Bacteriology. 191 (12): 3892–3900. doi:10.1128/jb.00975-08. PMC 2698385. PMID 19251852.
- 1 2 Howery KE, Clemmer KM, Şimşek E, Kim M, Rather PN (August 2015). Armitage JP (ed.). "Regulation of the Min Cell Division Inhibition Complex by the Rcs Phosphorelay in Proteus mirabilis". Journal of Bacteriology. 197 (15): 2499–2507. doi:10.1128/JB.00094-15. PMC 4518839. PMID 25986901.
- 1 2 Rather PN (August 2005). "Swarmer cell differentiation in Proteus mirabilis". Environmental Microbiology. 7 (8): 1065–1073. doi:10.1111/j.1462-2920.2005.00806.x. PMID 16011745.
- ↑ Morgenstein RM, Szostek B, Rather PN (September 2010). "Regulation of gene expression during swarmer cell differentiation in Proteus mirabilis". FEMS Microbiology Reviews. 34 (5): 753–763. doi:10.1111/j.1574-6976.2010.00229.x. PMID 20497230.
- ↑ Paustian TD (11 March 2007). "Proteus mirabilis and Urinary Tract Infection". Microbiology Textbook. University of Wisconsin - Madison. Archived from the original on 11 March 2007.
- ↑ O'Hara CM, Brenner FW, Miller JM (October 2000). "Classification, identification, and clinical significance of Proteus, Providencia, and Morganella". Clinical Microbiology Reviews. 13 (4): 534–46. doi:10.1128/CMR.13.4.534. PMC 88947. PMID 11023955.
- ↑ Mazumder SA, Gonzalez G. "Proteus Infections Medication". Medscape. Archived from the original on 26 October 2015. Retrieved 30 October 2015.
Further reading
- Esipov SE, Shapiro JA (1998). "Kinetic model of Proteus mirabilis swarm colony development". Journal of Mathematical Biology. 36 (3): 249–268. doi:10.1007/s002850050100. S2CID 59372912.
- Frénod E (2006). "Existence result for a model of Proteus mirabilis swarm". Differential and Integral Equations. 19 (6): 697–720. arXiv:math.FA/0702761. Bibcode:2007math......2761F.
- Gué M, Dupont V, Dufour A, Sire O (October 2001). "Bacterial swarming: a biochemical time-resolved FTIR-ATR study of Proteus mirabilis swarm-cell differentiation". Biochemistry. 40 (39): 11938–11945. doi:10.1021/bi010434m. PMID 11570895.
- Rauprich O, Matsushita M, Weijer CJ, Siegert F, Esipov SE, Shapiro JA (November 1996). "Periodic phenomena in Proteus mirabilis swarm colony development". Journal of Bacteriology. 178 (22): 6525–6538. doi:10.1128/jb.178.22.6525-6538.1996. PMC 178539. PMID 8932309.
- "Proteus mirabilis". NCBI Taxonomy Browser. 584. Archived from the original on 7 December 2021. Retrieved 21 February 2023.
External links
- Proteus Genome Projects Archived 27 September 2007 at the Wayback Machine from Genomes OnLine Database Archived 15 October 2011 at the Wayback Machine
- Type strain of Proteus mirabilis at BacDive – the Bacterial Diversity Metadatabase Archived 16 September 2016 at the Wayback Machine