Sapovirus
| Sapporo virus | |
|---|---|
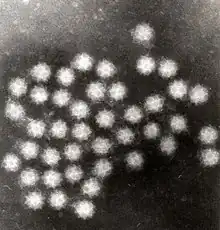 | |
| Transmission electron micrograph of Sapporo viruses | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Picornavirales |
| Family: | Caliciviridae |
| Genus: | Sapovirus |
| Species: | Sapporo virus |
Sapovirus is a genetically diverse genus of single-stranded positive-sense RNA, non-enveloped viruses within the family Caliciviridae.[1][2] Together with norovirus, sapoviruses are the most common cause of acute gastroenteritis (commonly called the "stomach flu" although it is not related to influenza) in humans and animals.[3][4] It is a monotypic taxon containing only one species, the Sapporo virus.[5]
Natural hosts for the virus are humans and swine. The virus is transmitted through oral/fecal contact. Sapovirus commonly occurs in children and infants and therefore is often spread in nurseries and daycares; however, it has also been found in long-term care facilities.[6] This could be due to a lack of personal hygiene and sanitation measures. Common symptoms include diarrhea and vomiting.[7] The sapovirus was initially discovered in an outbreak of gastroenteritis in an orphanage in Sapporo, Japan, in 1977.[8]
Viral classification
Structure and genome
Sapovirus is a non-enveloped, positive-sense, single-stranded RNA virus about 7.7kb in size. The virus has a 3'-end poly(A) tail but not a 5' cap. Sapovirus has an icosahedral structure that contains 180 subunits (T=3). The diameter of the capsid is between 27 and 40 nm.[9] Like other caliciviruses, the capsid of the sapovirus has round intends on its surface. However, its "Star of David" surface morphology distinguishes it from other caliciviruses.[3]
Sapovirus' genome is organized into two (possibly three) well-known open reading frames (ORFs). ORF1 encodes for nonstructural proteins and for VP1, the main capsid protein. VP1 has two standard domains, shell (S) and protruding (P). The S protein's function is to "form the scaffold around the nucleic acid", while the P protein is important in forming a "homodimer with the receptors".[10] ORF2 encodes for minor structural polyproteins, VP2. While there have been predictions of a third ORF (ORF3), there is no proof for what its function is.[3]
There have been at least 21 complete genomes for sapovirus analyzed and identified already, all of which can be classified into five categories (GI-GV), which can further be divided into different genetic clusters. Four of the five groups (GI, GII, GIV, GV) can infect humans and these four groups correspond to the four antigenically distinct strains of sapovirus: Sapporo, Houston, London, and Stockholm.[11] While there are at least 21 genotypes for this virus, new ones continue to be reported in America, Asia, and Europe.[12]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Sapovirus | Icosahedral | T=3 | Non-enveloped | Linear | Monopartite |
Transmission route and host susceptibility
Sapovirus is spread via the fecal–oral route. Infected individuals expel more than particles/gram of feces or vomit. Particles from the infected individual remain viable for years, and an infectious dose can be as few as 10 particles. Contamination of work surfaces, hands, etc. can cause a vast number of new infections. Infection may occur if the particles are inhaled, such as when the particles are aerosolized when those who are infected vomit, or when the toilet is flushed after an infected individual vomits. Other forms of transmission include the excessive handling of foods by an infected individual (this most commonly occurs in a restaurant setting), consumption of shellfish that lived in waters contaminated with infected fecal matter, and the ingestion of water that has been contaminated.[11]
Replication cycle
The exact replication cycle of sapovirus has not been determined; however, it is thought to have the same or similar cytoplasmic replication cycle that other caliciviruses display. The cytoplasmic replication cycle is as follows:
- Entry into the host cell is achieved by attachment to host receptors, which mediates endocytosis of the virus into the host cell.
- Uncoating and release of the viral genomic RNA into the cytoplasm.
- VPg is removed from the viral RNA, which is then translated into a processed ORF1 polyprotein to yield the replication proteins.
- Replication occurs in viral factories. A dsRNA genome is synthesized from the genomic ssRNA(+).
- The dsRNA genome is transcribed/replicated thereby providing viral mRNAs/new ssRNA(+) genomes.
- Subgenomic RNA translation gives rise to the capsid protein and VP2.
- Assembly of new virus particles and release by cell lysis."[9]
Infection
Symptoms and signs
After an incubation period of 1–4 days, signs of illness start to arise. Symptoms of sapovirus are very similar to those of norovirus. The most common symptoms are vomiting and diarrhea. However, additional symptoms may occur, including chills, nausea, headache, abdominal cramps, myalgia, and fever though it is very rare. While patients frequently start to show symptoms after the 1–4 day incubation period, there have been cases in which an individual is asymptomatic. Although the individual does not show symptoms, they are still capable of spreading the virus through the general mode of transmission, which is the oral-fecal route.[3]
Laboratory diagnosis
Nucleic acid detection methods
Reverse transcription-PCR (RT-PCR) is the most commonly used detection tool for sapovirus because of its broad reactivity, sensitivity, speed, and specificity. Because of the diversity of the sapovirus, hundreds of primers have been designed in order to specifically target and amplify RNA-dependent RNA polymerase. This can be used to "partially characterize the Sapovirus and investigate the similarity of the detected Sapovirus."[3]
Virus particle detection
"Sapoviruses are morphologically distinguishable from other gastroenteritis pathogens (e.g., norovirus, rotavirus, astrovirus, or adenovirus) by their typical "Star of David" surface morphology under the electron microscope. However, this has low sensitivity compared to nucleic acid detection methods."[3]
Antigen detection methods
Enzyme-linked immunosorbent assays (ELISA) have been used to detect human sapovirus from clinical samples. While ELISA can be used to detect human sapovirus antigens, it is not commonly used. The diversity of the many strains of sapovirus makes it difficult to detect the wide array of antigens that may be present. Because there are so many possible antigens, ELISA is not as accurate or as sensitive as the nucleic acid detection methods.[3]
Prevention
General sanitary hygiene is the most important method of preventing sapovirus. This can be done by thoroughly washing hands after using the restroom and before eating/preparing food. Contaminated surfaces should be cleaned with disinfectant and or solutions containing bleach. Other preventative measures include avoiding contact and sharing drinks/food with infected individuals.
In hospital settings
- Affected patients should be isolated and infected staff members should be suspended from work.
- Bed pan washers should be aware and educated so that they may be working with caution.
- If an extreme outbreak is to occur, "it may be necessary to close wards to new admissions."
- "Staff movement from affected to unaffected wards should be prohibited—group activities stopped and visits by children discouraged."[11]
Treatment
There is no specific medication for individuals infected with sapovirus. Sapovirus cannot be treated with antibiotics because it is not a bacterial infection. Treatments include symptom support such as rehydrating the individual.[11]
History
Using electron microscopy, the sapovirus was first seen in diarrheic stool samples from the United Kingdom in 1977 and was soon known as a gastroenteritis pathogen. While the virus was first seen in the United Kingdom, "the prototype strain of the genus Sapovirus was from another outbreak in Sapporo, Japan in 1982."[3] The first complete genome of sapovirus was interpreted from the Manchester strain in the United Kingdom in 1993. Formerly, sapoviruses were called "Sapporo-like viruses"; however, in 2002, they were changed to the species Sapporo virus, genus Sapovirus, in the family Caliciviridae. "Currently, the family Caliciviridae consists of five established genera: Sapovirus, Norovirus, Lagovirus, Vesivirus, and Nebovirus."[3]
Outbreaks
December 2013
One positive result for sapovirus infection was confirmed at Gisborne Hospital in New Zealand. Two additional cases were found in staff members and five additional patients were put into isolation. Hospital staff used precautionary measures by using personal protection when entering rooms.[8]
June 2007
Fifty five faculty members of a college in Taipei County had been diagnosed with sapovirus infection.
Long term care facilities 2002–2009
"Using data from the Oregon and Minnesota public health departments, researchers investigated 2161 gastroenteritis outbreaks between 2002 through 2009. Of these, 142 outbreaks (7 percent) were found to be norovirus-negative, and 93 of these were further tested for other gastrointestinal viruses including sapovirus, astrovirus, adenovirus, and rotavirus. Sapovirus was identified in 21 outbreaks (23 percent), with 66 percent of these occurring in long term care facilities. Close to half of these cases occurred in 2007 alone." The researchers further explained that while the proportion of sapovirus occurring in the long-term care facilities was high, it was likely an artifact of legally mandated outbreak reporting.[6]
Associated diseases
Norovirus is most commonly associated with sapovirus. Norovirus and sapovirus genomes are very closely related; the distinction between the two can only be made from the differences in their coding strategy and reading frames. Noroviruses, along with sapoviruses, are the most common cause of gastroenteritis and therefore show the same symptoms as each other.[14]
Astrovirus, like sapovirus, causes gastroenteritis in children and the elderly, especially those who are immunocompromised. While sapovirus has two ORFs, Astrovirus has three. Astrovirus also has 6 recombinant strains. Astrovirus replicates within the cytoplasm and propagates readily in the GI tract.[11]
Rotavirus, like norovirus, astrovirus, and sapovirus, causes gastroenteritis. Rotavirus, however, is much more lethal, causing 37% of deaths in children with diarrhea and 215,000 deaths worldwide.[15]
Animal viruses
Sapoviruses have been identified in bats, California sea lions, dogs, pigs and mink.[16][17]
References
- ↑ Vinjé, J; Estes, MK; Esteves, P; Green, KY; Katayama, K; Knowles, NJ; L'Homme, Y; Martella, V; Vennema, H; White, PA; ICTV Report Consortium (November 2019). "ICTV Virus Taxonomy Profile: Caliciviridae". The Journal of General Virology. 100 (11): 1469–1470. doi:10.1099/jgv.0.001332. PMC 7011698. PMID 31573467.
- ↑ "ICTV Report Caliciviridae". Archived from the original on 29 August 2021. Retrieved 13 August 2023.
- 1 2 3 4 5 6 7 8 9 Oka, Tomoichiro; Wang, Qiuhong; Katayama, Kazuhiko; Saif, Linda J. (1 January 2015). "Comprehensive Review of Human Sapoviruses". Clinical Microbiology Reviews. 28 (1): 32–53. doi:10.1128/CMR.00011-14. ISSN 0893-8512. PMC 4284302. PMID 25567221.
- ↑ Razizadeh, Mohammad Hossein; Khatami, Alireza; Zarei, Mohammad (8 October 2021). "Global molecular prevalence and genotype distribution of Sapovirus in children with gastrointestinal complications: A systematic review and meta‐analysis". Reviews in Medical Virology. 32 (3): e2302. doi:10.1002/rmv.2302. PMID 34626019. S2CID 238528430.
- ↑ "Virus Taxonomy: 2018b Release". International Committee on Taxonomy of Viruses (ICTV). March 2019. Archived from the original on 20 March 2020. Retrieved 1 May 2019.
- 1 2 Lee, Lore E.; Cebelinski, Elizabeth A.; Fuller, Candace; Keene, William E.; Smith, Kirk; Vinjé, Jan; Besser, John M. (2012). "Sapovirus Outbreaks in Long-Term Care Facilities, Oregon and Minnesota, USA, 2002–2009". Emerging Infectious Diseases. 18 (5): 873–876. doi:10.3201/eid1805.111843. PMC 3358050. PMID 22516204.
- ↑ "Virus Page". web.stanford.edu. Archived from the original on 19 November 2023. Retrieved 5 November 2017.
- 1 2 Harker-Smith, Marino (2 December 2013). "Sapovirus - New Zealand: (GI) hospital outbreak". Gisborne Herald. Archived from the original on 10 May 2023. Retrieved 2 November 2017 – via ProMED-mail.
- 1 2 "Viral Zone". ExPASy. Archived from the original on 15 June 2015. Retrieved 15 June 2015.
- ↑ Miyazaki, Naoyuki; Taylor, David W.; Hansman, Grant S.; Murata, Kazuyoshi (1 March 2016). "Antigenic and Cryo-Electron Microscopy Structure Analysis of a Chimeric Sapovirus Capsid". Journal of Virology. 90 (5): 2664–2675. doi:10.1128/JVI.02916-15. ISSN 0022-538X. PMC 4810723. PMID 26699644.
- 1 2 3 4 5 Barer, Mike (17 January 2011). "Caliciviruses and Astroviruses". In Greenwood, David (ed.). Medical Microbiology: A Guide to Microbial Infections: pathogenesis, immunity, laboratory diagnosis and control. pp. 579–586. ISBN 978-0702051197. Archived from the original on 6 May 2023. Retrieved 13 August 2023.
- ↑ Tsinda, Emmanuel Kagning; Malasao, Rungnapa; Furuse, Yuki; Gilman, Robert H.; Liu, Xiaofang; Apaza, Sonia; Espetia, Susan; Cama, Vitaliano; Oshitani, Hitoshi (26 October 2017). "Complete Coding Genome Sequences of Uncommon GII.8 Sapovirus Strains Identified in Diarrhea Samples Collected from Peruvian Children". Genome Announcements. 5 (43): e01137–17. doi:10.1128/genomeA.01137-17. ISSN 2169-8287. PMC 5658493. PMID 29074655.
- ↑ Makhaola, Kgomotso; Moyo, Sikhulile; Kebaabetswe, Lemme P. (27 April 2020). "Distribution and Genetic Variability of Sapoviruses in Africa". Viruses. 12 (5): 490. doi:10.3390/v12050490. ISSN 1999-4915. Retrieved 18 November 2023.
- ↑ Farkas, T.; Zhong, W. M.; Jing, Y.; Huang, P. W.; Espinosa, S. M.; Martinez, N.; Morrow, A. L.; Ruiz-Palacios, G. M.; Pickering, L. K. (1 July 2004). "Genetic diversity among sapoviruses". Archives of Virology. 149 (7): 1309–1323. doi:10.1007/s00705-004-0296-9. ISSN 0304-8608. PMID 15221533. S2CID 35990669.
- ↑ Tate, Jacqueline E.; Burton, Anthony H.; Boschi-Pinto, Cynthia; Parashar, Umesh D.; World Health Organization–Coordinated Global Rotavirus Surveillance Network (1 May 2016). "Global, Regional, and National Estimates of Rotavirus Mortality in Children". Clinical Infectious Diseases. 62 Suppl 2: S96–S105. doi:10.1093/cid/civ1013. ISSN 1537-6591. PMID 27059362.
- ↑ Meng XJ (January 2012). "Emerging and Re-emerging Swine Viruses". Transboundary and Emerging Diseases. 59: 85–102. doi:10.1111/j.1865-1682.2011.01291.x. PMID 22225855.
- ↑ Tse, H.; Chan, W. M.; Li, K. S.; Lau, S. K.; Woo, P. C.; Yuen, K. Y. (2012). "Discovery and Genomic Characterization of a Novel Bat Sapovirus with Unusual Genomic Features and Phylogenetic Position". PLOS ONE. 7 (4): e34987. doi:10.1371/journal.pone.0034987. PMC 3325917. PMID 22514697.
External links
- ICTV Report: Caliciviridae Archived 29 August 2021 at the Wayback Machine
- Viralzone: Sapovirus Archived 4 March 2016 at the Wayback Machine