HLA-B70
HLA-B70 (B70) is an HLA-B serotype. The serotype identifies certain B*15 gene-allele protein products of HLA-B.[1] B70 is one of many split antigens of the broad antigen, B15.
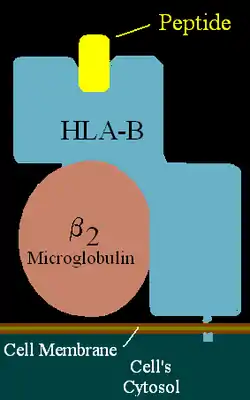 | ||
| HLA-B (alpha)-β2MG with bound peptide | ||
major histocompatibility complex (human), class I, B70 | ||
| Alleles | *1503, *1509, *1510, *1518, *1529, *1537 | |
| Structure (See HLA-B) | ||
| Shared data | ||
| Locus | chr.6 6p21.31 | |
Serotype
| B*15 | B15 | B62 | B63 | B70 | B71 | B72 | B75 | B76 | B77 | Sample |
| allele | % | % | % | % | % | % | % | % | % | size (N) |
| *1503 | 1 | 61 | 10 | 2569 | ||||||
| *1509 | 53 | 65 | ||||||||
| *1510 | 1 | 49 | 2 | 2 | 1 | 1204 | ||||
| *1518 | 51 | 3 | 552 | |||||||
| *1529 | 19 | 21 | ||||||||
| *1537 | 30 | 34 | ||||||||
| Alleles link-out to IMGT/HLA Databease at EBI | ||||||||||
Alleles
| freq | ||
| ref. | Population | (%) |
| [3] | Burkina Faso Rimaibe | 13.8 |
| Guinea Bissau | 10.8 | |
| Cameroon Pygmy Baka | 10.0 | |
| South African Natal Zulu | 9.5 | |
| Kenya Luo | 8.9 | |
| Ivory Coast Akan Adiopodoume | 8.0 | |
| Kenya Nandi | 7.9 | |
| Cameroon Sawa | 7.7 | |
| Zimbabwe Harare Shona | 7.7 | |
| USA African America | 7.3 | |
| Burkina Faso Fulani | 7.1 | |
| Mali Bandiagara | 6.9 | |
| Senegal Niokholo Mandenka | 6.9 | |
| Cape Verde SE Islands | 6.5 | |
| Kenya | 6.3 | |
| Uganda Kampala | 6.2 | |
| Sudanese | 6.0 | |
| Zambia Lusaka | 5.7 | |
| Cameroon Yaounde | 4.9 | |
| Cape Verde NW Islands | 4.8 | |
| Oman | 4.7 | |
| Cameroon Bamileke | 4.5 | |
| Tunisia Tunis | 4.5 | |
| Cameroon Beti | 3.7 | |
| Brazil Belo Horizonte | 3.2 | |
| Azores Terceira Island | 2.4 | |
| Madeira | 2.2 | |
| Saudi Arabia Guraiat and Hail | 2.0 | |
| Tunisia | 2.0 | |
| United Arab Emerates | 1.5 | |
| Brazil | 1.4 | |
| Cuban White | 1.4 | |
| Morocco Nador Metalsa | 1.4 | |
| Mexico Mestizos | 1.2 | |
| Italy Bergamo | 1.1 | |
| Portugal North | 1.1 | |
| American Samoa | 1.0 | |
| Iran Baloch | 1.0 | |
| Mexico Guadalajara Mestizos (2) | 1.0 | |
| Azores Central Islands | 0.9 | |
| Israel Ashk. and Non-Ashk. Jews | 0.9 | |
| Taiwan Hakka | 0.9 | |
| Croatia | 0.7 |
| freq | ||
| ref. | Population | (%) |
| [3] | Burkina Faso Fulani | 4.1 |
| India West Coast Parsis | 3.0 | |
| Spain Eastern Andalusia Gipsy | 3.0 | |
| Mexico Mestizos | 2.4 | |
| Italy South Campania | 1.2 | |
| Burkina Faso Rimaibe | 1.1 | |
| China Inner Mongolia | 1.0 | |
| Mali Bandiagara | 0.7 | |
| Czech Republic | 0.5 |
| freq | ||
| ref. | Population | (%) |
| [3] | South African Natal Zulu | 8.5 |
| Zimbabwe Harare Shona | 8.4 | |
| Ivory Coast Akan Adiopodoume | 5.7 | |
| Kenya | 5.2 | |
| Zambia Lusaka | 4.6 | |
| Cameroon Sawa | 3.8 | |
| Guinea Bissau | 3.8 | |
| Kenya Luo | 3.6 | |
| Senegal Niokholo Mandenka | 3.2 | |
| Cameroon Beti | 2.8 | |
| USA African Americans (3) | 2.7 | |
| Uganda Kampala | 2.5 | |
| Mexico Mestizos | 2.4 | |
| Mali Bandiagara | 2.2 | |
| Kenya Nandi | 1.7 | |
| Brazil Belo Horizonte | 1.6 | |
| Cameroon Bamileke | 1.3 | |
| Cuban Mulatto | 1.2 | |
| Cameroon Yaounde | 1.1 | |
| Iran Baloch | 1.0 | |
| Sudanese | 1.0 | |
| Tunisia | 1.0 | |
| Brazil Terena | 0.9 | |
| Cape Verde NW Islands | 0.8 | |
| United Arab Emerates | 0.8 | |
| USA North American Natives | 0.8 | |
| Guadalajara Mestizos (2) | 0.5 |
| freq | ||
| ref. | Population | (%) |
| [3] | South Africa Natal Tamil | 4.1 |
| Autonomous Tibetans | 3.8 | |
| India Mumbai Marathas | 3.7 | |
| China North Han | 2.9 | |
| USA Hawaii Okinawa | 2.9 | |
| China Guangzhou Han | 2.4 | |
| India Andhra Pradesh Golla | 2.4 | |
| China Beijing | 2.3 | |
| Taiwan Hakka | 1.8 | |
| Japan | 1.7 | |
| India North Delhi | 1.6 | |
| China Inner Mongolia | 1.5 | |
| China Qinghai Hui | 1.4 | |
| Czech Republic | 1.4 | |
| China Yunnan Nu | 1.3 | |
| Shijiazhuang Tianjian Han | 1.1 | |
| Ivory Coast Akan Adiopodoume | 1.1 | |
| Senegal Niokholo Mandenka | 1.1 | |
| Zambia Lusaka | 1.1 | |
| Portugal Centre | 1.0 | |
| Singapore Javanese Indonesians | 1.0 | |
| Thailand (3) | 1.0 | |
| South Korea (3) | 0.9 | |
| Cape Verde Islands | 0.8 | |
| France South East | 0.8 | |
| Guinea Bissau | 0.8 | |
| China South Han | 0.7 | |
| China Yunnan Lisu | 0.7 | |
| Russia Tuva (2) | 0.6 | |
| Singapore Chinese Han | 0.6 | |
| Spain Eastern Andalusia | 0.6 |
References
- Marsh, S. G.; Albert, E. D.; Bodmer, W. F.; Bontrop, R. E.; Dupont, B.; Erlich, H. A.; Fernández-Viña, M.; Geraghty, D. E.; Holdsworth, R.; Hurley, C. K.; Lau, M.; Lee, K. W.; Mach, B.; Maiers, M.; Mayr, W. R.; Müller, C. R.; Parham, P.; Petersdorf, E. W.; Sasazuki, T.; Strominger, J. L.; Svejgaard, A.; Terasaki, P. I.; Tiercy, J. M.; Trowsdale, J. (2010). "Nomenclature for factors of the HLA system, 2010". Tissue Antigens. 75 (4): 291–455. doi:10.1111/j.1399-0039.2010.01466.x. PMC 2848993. PMID 20356336.
- derived from IMGT/HLA
- Middleton D, Menchaca L, Rood H, Komerofsky R (2003). "New allele frequency database: http://www.allelefrequencies.net". Tissue Antigens. 61 (5): 403–7. doi:10.1034/j.1399-0039.2003.00062.x. PMID 12753660.
{{cite journal}}: External link in|title=
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.