Amelogenesis imperfecta
| Amelogenesis imperfecta | |
|---|---|
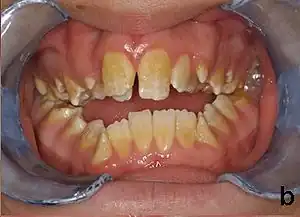 | |
| Amelogenesis imperfecta, hypoplastic type. Note the association of pitted enamel and open bite. | |
Amelogenesis imperfecta (AI) is a congenital disorder which presents with a rare abnormal formation of the enamel[1] or external layer of the crown of teeth, unrelated to any systemic or generalized conditions.[2] Enamel is composed mostly of mineral, that is formed and regulated by the proteins in it. Amelogenesis imperfecta is due to the malfunction of the proteins in the enamel (ameloblastin, enamelin, tuftelin and amelogenin) as a result of abnormal enamel formation via amelogenesis.[3]
People afflicted with amelogenesis imperfecta may have teeth with abnormal color: yellow, brown or grey; this disorder can afflict any number of teeth of both dentitions. Enamel hypoplasia manifests in a variety of ways depending on the type of AI an individual has (see below), with pitting and plane-form defects common.[4] The teeth have a higher risk for dental cavities and are hypersensitive to temperature changes as well as rapid attrition, excessive calculus deposition, and gingival hyperplasia.[5] The earliest known case of AI is in an extinct hominid species called Paranthropus robustus, with over a third of individuals displaying this condition.[6]
Symptoms and signs

The clinical presentation of Amelogenesis imperfecta is as follows:[7]
- Abnormal jaw morphology
- Multiple unerupted teeth
- Taurodontia
- Brown discoloration teeth
- Anterior open bite
- Impaired mastication
Genetics
Several gene expression is needed for enamel formation where the relevant matrix proteins & proteinases are transcribed for regular crystal growth & enamel mineralization.
Mutations in the AMELX,[8] ENAM,[9] MMP20,[10] KLK-4,[11] FAM83H,[12] WDR72,[13] C4orf26,[14] SLC24A4[15][16] LAMB3[17] and ITGB6[18] genes have been found to cause amelogenesis imperfecta (non-syndromic form). AMELX and ENAM encode extracellular matrix proteins of the developing tooth enamel and KLK-4 and MMP20 encode proteases that help degrade organic matter from the enamel matrix during the maturation stage of amelogenesis. SLC24A4 encodes a calcium transporter that mediates calcium transport to developing enamel during tooth development. Less is known about the function of other genes implicated in amelogenesis imperfecta.
Researchers expect that mutations in further genes are likely to be identified as causes of amelogenesis imperfecta. Types include:
| Type | OMIM | Gene | Locus |
|---|---|---|---|
| AI1B | 104500 | ENAM | 4q21 |
| AI1C | 204650 | ENAM | 4q21 |
| AI2A1 | 204700 | KLK4 | 19q13.4 |
| AI2A2 | 612529 | MMP20 | 11q22.3-q23 |
| AI2A3 | 613211 | WDR72 | 15q21.3 |
| AI2A4 | 614832 | ODAPH | 4q21.1 |
| AI2A5 | 609840 | SLC24A4 | 14q32.12 |
| AI3 | 130900 | FAM83H | 8q24.3 |
| AIH1 | 301200 | AMELX | Xp22.3-p22.1 |
| AIGFS | 614253 | FAM20A | 17q24.2 |
Amelogenesis imperfecta can have different inheritance patterns depending on the gene that is altered. Mutations in the ENAM gene are the most frequent known cause and are most commonly inherited in an autosomal dominant pattern. This type of inheritance means one copy of the altered gene in each cell is sufficient to cause the disorder.
Amelogenesis imperfecta is also inherited in an autosomal recessive pattern; this form of the disorder can result from mutations in the ENAM, MMP20, KLK4, FAM20A, C4orf26 or SLC24A4 genes. Autosomal recessive inheritance means two copies of the gene in each cell are altered.
About 5% of amelogenesis imperfecta cases are caused by mutations in the AMELX gene and are inherited in an X-linked pattern. A condition is considered X-linked if the mutated gene that causes the disorder is located on the X chromosome, one of the two sex chromosomes. In most cases, males with an X-linked form of this condition experience more severe dental abnormalities than affected females. Recent genetic studies suggest that the cause of a significant proportion of amelogenesis imperfecta cases remains to be discovered.
Diagnosis
AI can be classified according to their clinical appearances:[19]
- Type 1 - Hypoplastic
- Enamel of abnormal thickness due to malfunction in enamel matrix formation. Enamel is very thin but hard & translucent, and may have random pits & grooves. Condition is of autosomal dominant, autosomal recessive, or x-linked pattern. Enamel differs in appearance from dentine radiographically as normal functional enamel.[20]
- Type 2 - Hypomaturation
- Enamel has sound thickness, with a pitted appearance. It is less hard compared to normal enamel, and are prone to rapid wear, although not as intense as Type 3 AI. Condition is of autosomal dominant, autosomal recessive, or x-linked pattern. Enamel appears to be comparable to dentine in its radiodensity on radiographs.
- Type 3 - Hypocalcified
- Enamel defect due to malfunction of enamel calcification, therefore enamel is of normal thickness but is extremely brittle, with an opaque/chalky presentation. Teeth are prone to staining and rapid wear, exposing dentine. Condition is of autosomal dominant and autosomal recessive pattern. Enamel appears less radioopaque compared to dentine on radiographs.
- Type 4 - Hypomature hypoplastic enamel with taurodontism
- Enamel has a variation in appearance, with mixed features from Type 1 and Type 2 AI. All Type 4 AI has taurodontism in common. Condition is of autosomal dominant pattern. Other common features may include an anterior open bite,[21] taurodontism, sensitivity of teeth.
Differential diagnosis would include dental fluorosis, molar-incisor hypomineralization, chronological disorders of tooth development.[22]
Treatment

Preventive and restorative dental care is very important as well as considerations for esthetic issues since the crown are yellow from exposure of dentin due to enamel loss.[5] The main objectives of treatment is pain relief, preserving patient's remaining dentition, and to treat and preserve the patient's occlusal vertical height.[20]
Many factors are to be considered to decide on treatment options such as the classification and severity of AI, the patient's social history, clinical findings etc. There are many classifications of AI but the general management of this condition is similar.
Full-coverage crowns are sometimes being used to compensate for the abraded enamel in adults, tackling the sensitivity the patient experiences. Usually stainless steel crowns are used in children which may be replaced by porcelain once they reach adulthood.[23] These aid with maintaining occlusal vertical dimension.
Aesthetics may be addressed via placement of composite or porcelain veneers, depending on patient factors e.g. age. If the patient has primary or mixed dentition, lab-made composite veneers may be provided temporarily, to be replaced by permanent porcelain veneers once the patient has stabilized permanent dentition. The patient's oral hygiene and diet should be controlled as well as they play a factor in the success of retaining future restorations.
In the worst-case scenario, the teeth may have to be extracted and implants or dentures are required. Loss of nerves in the affected teeth may occur.
Epidemiology
The exact incidence of amelogenesis imperfecta is uncertain. Estimates vary widely, from 1 in 700 people in northern Sweden to 1 in 14,000 people in the United States.[24] The prevalence of amelogenesis imperfecta in non-human animals has not been explored, however its presence has been noted.[25]
This condition is neither caused by nor the equivalent of dental fluorosis. A manifestation of amelogenesis imperfecta known as "snow capping" is confined to the outer prismless enamel layer. It may superficially resemble dental fluorosis, and indeed "snow capping" may be used as a descriptive term in some incidents of dental fluorosis.[26][27]
References
- ↑ Slootweg PJ (2007). Dental pathology: a practical introduction. Springer Science & Business Media. pp. 19–. ISBN 978-3-540-71690-7. Archived from the original on 29 June 2011. Retrieved 28 December 2010.
- ↑ Kida M, Ariga T, Shirakawa T, Oguchi H, Sakiyama Y (November 2002). "Autosomal-dominant hypoplastic form of amelogenesis imperfecta caused by an enamelin gene mutation at the exon-intron boundary". Journal of Dental Research. 81 (11): 738–42. doi:10.1177/154405910208101103. PMID 12407086.
- ↑ Smith CE, Murillo G, Brookes SJ, Poulter JA, Silva S, Kirkham J, Inglehearn CF, Mighell AJ (August 2016). "Deletion of amelotin exons 3-6 is associated with amelogenesis imperfecta". Human Molecular Genetics. 25 (16): 3578–3587. doi:10.1093/hmg/ddw203. PMC 5179951. PMID 27412008.
- ↑ Crawford PJ, Aldred M, Bloch-Zupan A (April 2007). "Amelogenesis imperfecta". Orphanet Journal of Rare Diseases. 2 (1): 17. doi:10.1186/1750-1172-2-17. PMC 1853073. PMID 17408482.
- 1 2 American Academy of Pediatric Dentistry, Guideline on Dental Management of Heritable Dental Developmental Anomalies, 2013, http://www.aapd.org/media/Policies_Guidelines/G_OHCHeritable.pdf Archived 2019-03-27 at the Wayback Machine
- ↑ Towle, Ian; Irish, Joel D. (2019). "A probable genetic origin for pitting enamel hypoplasia on the molars of Paranthropus robustus" (PDF). Journal of Human Evolution. 129: 54–61. doi:10.1016/j.jhevol.2019.01.002. PMID 30904040. Archived (PDF) from the original on 2020-08-06. Retrieved 2020-10-19.
- ↑ "Amelogenesis imperfecta | Genetic and Rare Diseases Information Center (GARD) – an NCATS Program". rarediseases.info.nih.gov. Archived from the original on 2 July 2021. Retrieved 15 August 2021.
- ↑ Lagerström M, Dahl N, Nakahori Y, Nakagome Y, Bäckman B, Landegren U, Pettersson U (August 1991). "A deletion in the amelogenin gene (AMG) causes X-linked amelogenesis imperfecta (AIH1)". Genomics. 10 (4): 971–5. doi:10.1016/0888-7543(91)90187-j. PMID 1916828.
- ↑ Rajpar MH, Harley K, Laing C, Davies RM, Dixon MJ (August 2001). "Mutation of the gene encoding the enamel-specific protein, enamelin, causes autosomal-dominant amelogenesis imperfecta". Human Molecular Genetics. 10 (16): 1673–7. doi:10.1093/hmg/10.16.1673. PMID 11487571.
- ↑ Kim JW, Simmer JP, Hart TC, Hart PS, Ramaswami MD, Bartlett JD, Hu JC (March 2005). "MMP-20 mutation in autosomal recessive pigmented hypomaturation amelogenesis imperfecta". Journal of Medical Genetics. 42 (3): 271–5. doi:10.1136/jmg.2004.024505. PMC 1736010. PMID 15744043.
- ↑ Hart PS, Hart TC, Michalec MD, Ryu OH, Simmons D, Hong S, Wright JT (July 2004). "Mutation in kallikrein 4 causes autosomal recessive hypomaturation amelogenesis imperfecta". Journal of Medical Genetics. 41 (7): 545–9. doi:10.1136/jmg.2003.017657. PMC 1735847. PMID 15235027.
- ↑ Kim JW, Lee SK, Lee ZH, Park JC, Lee KE, Lee MH, Park JT, Seo BM, Hu JC, Simmer JP (February 2008). "FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta". American Journal of Human Genetics. 82 (2): 489–94. doi:10.1016/j.ajhg.2007.09.020. PMC 2427219. PMID 18252228.
- ↑ El-Sayed W, Parry DA, Shore RC, Ahmed M, Jafri H, Rashid Y, et al. (November 2009). "Mutations in the beta propeller WDR72 cause autosomal-recessive hypomaturation amelogenesis imperfecta". American Journal of Human Genetics. 85 (5): 699–705. doi:10.1016/j.ajhg.2009.09.014. PMC 2775821. PMID 19853237.
- ↑ Parry DA, Brookes SJ, Logan CV, Poulter JA, El-Sayed W, Al-Bahlani S, Al Harasi S, Sayed J, el Raïf M, Shore RC, Dashash M, Barron M, Morgan JE, Carr IM, Taylor GR, Johnson CA, Aldred MJ, Dixon MJ, Wright JT, Kirkham J, Inglehearn CF, Mighell AJ (September 2012). "Mutations in C4orf26, encoding a peptide with in vitro hydroxyapatite crystal nucleation and growth activity, cause amelogenesis imperfecta". American Journal of Human Genetics. 91 (3): 565–71. doi:10.1016/j.ajhg.2012.07.020. PMC 3511980. PMID 22901946.
- ↑ Parry DA, Poulter JA, Logan CV, Brookes SJ, Jafri H, Ferguson CH, et al. (February 2013). "Identification of mutations in SLC24A4, encoding a potassium-dependent sodium/calcium exchanger, as a cause of amelogenesis imperfecta". American Journal of Human Genetics. 92 (2): 307–12. doi:10.1016/j.ajhg.2013.01.003. PMC 3567274. PMID 23375655.
- ↑ Herzog CR, Reid BM, Seymen F, Koruyucu M, Tuna EB, Simmer JP, Hu JC (February 2015). "Hypomaturation amelogenesis imperfecta caused by a novel SLC24A4 mutation". Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology. 119 (2): e77–81. doi:10.1016/j.oooo.2014.09.003. PMC 4291293. PMID 25442250.
- ↑ Poulter JA, El-Sayed W, Shore RC, Kirkham J, Inglehearn CF, Mighell AJ (January 2014). "Whole-exome sequencing, without prior linkage, identifies a mutation in LAMB3 as a cause of dominant hypoplastic amelogenesis imperfecta". European Journal of Human Genetics. 22 (1): 132–5. doi:10.1038/ejhg.2013.76. PMC 3865405. PMID 23632796.
- ↑ Wang SK, Choi M, Richardson AS, Reid BM, Lin BP, Wang SJ, Kim JW, Simmer JP, Hu JC (April 2014). "ITGB6 loss-of-function mutations cause autosomal recessive amelogenesis imperfecta". Human Molecular Genetics. 23 (8): 2157–63. doi:10.1093/hmg/ddt611. PMC 3959820. PMID 24305999.
- ↑ Fonseca RB, Sobrinho LC, Neto AJ, Soares da Mota A, Soares CJ (2006). "Enamel hypoplasia or amelogenesis imperfecta – a restorative approach". Brazilian Journal of Oral Sciences. 5 (16): 941–3.
- 1 2 Visram S, McKaig S (December 2006). "Amelogenesis imperfecta--clinical presentation and management: a case report". Dental Update. 33 (10): 612–4, 616. doi:10.12968/denu.2006.33.10.612. PMID 17209536.
- ↑ Bouvier D, Duprez JP, Bois D (1996). "Rehabilitation of young patients with amelogenesis imperfecta: a report of two cases". ASDC Journal of Dentistry for Children. 63 (6): 443–7. PMID 9017180.
- ↑ Crawford PJ, Aldred M, Bloch-Zupan A (April 2007). "Amelogenesis imperfecta". Orphanet Journal of Rare Diseases. 2: 17. doi:10.1186/1750-1172-2-17. PMC 1853073. PMID 17408482.
- ↑ Illustrated Dental Embryology, Histology, and Anatomy, Bath-Balogh and Fehrenbach, Elsevier, 2011, page 64
- ↑ Hoppenreijs TJ, Voorsmit RA, Freihofer HP (August 1998). "Open bite deformity in amelogenesis imperfecta. Part 1: An analysis of contributory factors and implications for treatment". Journal of Cranio-Maxillo-Facial Surgery. 26 (4): 260–6. doi:10.1016/s1010-5182(98)80023-1. PMID 9777506.
- ↑ Towle I, Irish JD, De Groote I (April 2018). "Amelogenesis imperfecta in the dentition of a wild chimpanzee" (PDF). Journal of Medical Primatology. 47 (2): 117–119. doi:10.1111/jmp.12323. PMID 29112236. S2CID 3801451. Archived (PDF) from the original on 2019-04-28. Retrieved 2020-10-19.
- ↑ Chaudhary M, Dixit S, Singh A, Kunte S (July 2009). "Amelogenesis imperfecta: Report of a case and review of literature". Journal of Oral and Maxillofacial Pathology. 13 (2): 70–7. doi:10.4103/0973-029X.57673. PMC 3162864. PMID 21887005.
- ↑ Hu JC, Chan HC, Simmer SG, Seymen F, Richardson AS, Hu Y, Milkovich RN, Estrella NM, Yildirim M, Bayram M, Chen CF, Simmer JP (2012). "Amelogenesis imperfecta in two families with defined AMELX deletions in ARHGAP6". PLOS ONE. 7 (12): e52052. Bibcode:2012PLoSO...752052H. doi:10.1371/journal.pone.0052052. PMC 3522662. PMID 23251683.
Further reading
- Winter GB, Brook AH (January 1975). "Enamel hypoplasia and anomalies of the enamel". Dental Clinics of North America. 19 (1): 3–24. PMID 162891.
- Simmer JP, Hu JC (September 2001). "Dental enamel formation and its impact on clinical dentistry". Journal of Dental Education. 65 (9): 896–905. doi:10.1002/j.0022-0337.2001.65.9.tb03438.x. PMID 11569606.
- Aldred MJ, Savarirayan R, Crawford PJ (January 2003). "Amelogenesis imperfecta: a classification and catalogue for the 21st century". Oral Diseases. 9 (1): 19–23. doi:10.1034/j.1601-0825.2003.00843.x. PMID 12617253.
- Nusier M, Yassin O, Hart TC, Samimi A, Wright JT (February 2004). "Phenotypic diversity and revision of the nomenclature for autosomal recessive amelogenesis imperfecta". Oral Surgery, Oral Medicine, Oral Pathology, Oral Radiology, and Endodontics. 97 (2): 220–30. doi:10.1016/j.tripleo.2003.08.007. PMID 14970781.
- Stephanopoulos G, Garefalaki ME, Lyroudia K (December 2005). "Genes and related proteins involved in amelogenesis imperfecta". Journal of Dental Research. 84 (12): 1117–26. doi:10.1177/154405910508401206. PMID 16304440. S2CID 17693799.
External links
| Classification | |
|---|---|
| External resources |