Maitotoxin
Maitotoxin (or MTX) is an extremely potent biotoxin produced by Gambierdiscus toxicus, a dinoflagellate species. Maitotoxin has been shown to be more than one hundred thousand times more potent than VX nerve agent.[1] Maitotoxin is so potent that it has been demonstrated that an intraperitoneal injection of 130 ng/kg was lethal in mice.[2] Maitotoxin was named from the ciguateric fish Ctenochaetus striatus—called "maito" in Tahiti—from which maitotoxin was isolated for the first time. It was later shown that maitotoxin is actually produced by the dinoflagellate Gambierdiscus toxicus.
 | |
 | |
| Names | |
|---|---|
| Preferred IUPAC name
Disodium (12S,14aR,15aS,16aR,17aS,18Z,110aR,111aS,112aR,113aS,114aR,116R,117R,118aS,119aR,121aS,122aR,123aS,124aR,125aS,126aR,127aS,22S,24aR,25aS,26aR,27aS,28aR,29aS,211R,212R,213aR,214S,214aS,215aR,217aS,218aR,219aS,32R,33R,34aS,36S,37R,38R,38aS,5R,7R,82S,83R,84aS,86R,87R,88R,88aS,92R,93R,94R,94aS,95aS,96aR,97aS,98R,99R,910S,911aR,912aS,913aR,914R,914aR,11S,12R,132S,133R,134S,134aS,135aR,136aS,137aR,138S,138aS,1310S,1311R,1312aR,1313aS,1314aR,1315aS,1317R,1317aR)-12-[(1S,2R,4R,5S)-1,2-dihydroxy-4,5-dimethyloct-7-en-1-yl]-117,211,214,33,37,38,5,7,83,87,88,93,94,98,914,11,12,133,134,138,1311,1317-docosahydroxy-14a,15a,16a,114a,116,119a,121a,122a,25a,27a,29a,214a,217a,1313a,1315a-pentadecamethyl-132-[(2R,3R,4R,7S,8R,9R,11R,13E)-3,8,11,15-tetrahydroxy-4,9,13-trimethyl-12-methylidene-7-(sulfonatooxy)pentadec-13-en-2-yl]-13,14,14a,15a,16,16a,17a,110,110a,111a,112,112a,113a,114,114a,116,117,118,118a,119a,120,121,121a,122a,123,123a,124a,125,125a,126a,127,127a,22,23,24,24a,25a,26,26a,27a,28,28a,29a,210,211,212,213a,214,214a,215a,216,217,217a,218a,219,219a,32,33,34,34a,36,37,38,38a,82,83,84,84a,86,87,88,88a,93,94,94a,95a,96,96a,97a,98,99,910,911a,912,912a,913a,914,914a,133,134,134a,135a,136,136a,137a,138,138a,1310,1311,1312,1312a,1313a,1314,1314a,1315a,1316,1317,1317a-octahectahydro-12H,92H,132H-1(16)-pyrano[2′′′ ′,3′′′ ′:5′′′,6′′′]pyrano[2′′′,3′′′:6′′,7′′]oxepino[2′′,3′′:5′,6′]pyrano[2′,3′:5,6]pyrano[3,2-b]pyrano[2′′′,3′′′:5′′,6′′]pyrano[2′′,3′′:5′,6′]pyrano[2′,3′:5,6]pyrano[2,3-g]oxocina-2(2,12)-bis(pyrano[2′′,3′′:5,6]pyrano[2′,3′:5,6]pyrano)[3,2-b:2′,3′-f]oxepina-13(10)-pyrano[3,2-b]pyrano[2′′′,3′′′:5′′,6′′]pyrano[2′′,3′′:5′,6′]pyrano[2′,3′:5,6]pyrano[2,3-f]oxepina-9(2,10)-dipyrano[2,3-e:2′,3′-e′]pyrano[3,2-b:5,6-b′]dipyrana-3,8(2,6)-bis(pyrano[3,2-b]pyrana)tridecaphan-99-yl sulfate | |
| Identifiers | |
3D model (JSmol) |
|
| ChEMBL | |
| ChemSpider | |
| ECHA InfoCard | 100.227.039 |
| KEGG | |
PubChem CID |
|
| UNII | |
CompTox Dashboard (EPA) |
|
| |
| |
| Properties | |
| C164H256O68S2Na2 | |
| Molar mass | 3422 g/mol |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa).
Infobox references | |
Mechanism of toxicity
Maitotoxin activates extracellular calcium channels, leading to an increase in levels of cytosolic Ca2+ ions.[3] The exact molecular target of maitotoxin is unknown, but it has been suggested that maitotoxin binds to the plasma membrane Ca2+ ATPase (PMCA) and turns it into an ion channel, similar to how palytoxin turns the Na+/K+-ATPase into an ion channel.[4] Ultimately, a necroptosis cascade is activated, resulting in membrane blebbing and eventually cell lysis.[5] Maitotoxin can indirectly activate calcium-binding proteases calpain-1 and calpain-2, contributing to necrosis.[6] The toxicity of maitotoxin in mice is the highest for nonprotein toxins: the LD50 is 50 ng/kg.[7]
Molecular structure
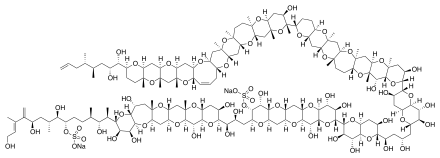
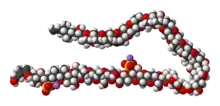
The molecule itself is a system of 32 fused rings. It resembles large fatty acid chains and it is notable because it is one of the largest and most complex non-protein, non-polysaccharide molecules produced by any organism. Maitotoxin includes 32 ether rings, 22 methyl groups, 28 hydroxyl groups, and 2 sulfuric acid esters and has an amphipathic structure.[8][9][10] Its structure was established through analysis using nuclear magnetic resonance at Tohoku University, Harvard University and the University of Tokyo in combination with mass spectrometry, and synthetic chemical methods. However, Andrew Gallimore and Jonathan Spencer have questioned the structure of maitotoxin at a single ring-junction (the J–K junction), based purely on biosynthetic considerations and their general model for marine polyether biogenesis.[11] K. C. Nicolaou and Michael Frederick argue that despite this biosynthetic argument, the originally proposed structure could still be correct. [12] The controversy has yet to be resolved.
Biosynthesis
The molecule is produced in nature via a polyketide synthase pathway.[11]
Total synthesis
Since 1996 the Nicolaou research group is involved in an effort to synthesise the molecule via total synthesis[13][14][15][16] although as of 2015 the project is on hold due to lack of funding.[17]
See also
References
- "MSDS: VX". www.ilpi.com. Retrieved 2022-09-29.
- Yokoyama, A; et al. (1988). "Some Chemical Properties of Maitotoxin, a Putative Calcium Channel Agonist Isolated from a Marine Dinoflagellate". J. Biochem. 104 (2): 184–187. doi:10.1093/oxfordjournals.jbchem.a122438. PMID 3182760.
- Ohizumi, Y; Yasumoto, T (1983). "Contraction and increase in tissue calcium content induced by maitotoxin, the most potent known marine toxin, in intestinal smooth muscle". British Journal of Pharmacology. 79 (1): 3–5. doi:10.1111/j.1476-5381.1983.tb10485.x. PMC 2044839. PMID 6871549.
- Sinkins, W. G; Estacion, M; Prasad, V; Goel, M; Shull, G. E; Kunze, D. L; Schilling, W. P (2009). "Maitotoxin converts the plasmalemmal Ca2+ pump into a Ca2+-permeable nonselective cation channel". American Journal of Physiology. Cell Physiology. 297 (6): C1533–43. doi:10.1152/ajpcell.00252.2009. PMC 2793065. PMID 19794142.
- Estacion, M & Schilling, WP (2001). "Maitotoxin-induced membrane blebbing and cell death in bovine aortic endothelial cells". BMC Physiology. 1: 2. doi:10.1186/1472-6793-1-2. PMC 32181. PMID 11231888.
- Wang, K.; et al. (1996). "Maitotoxin induces calpain activation in SH-SY5Y neuroblastoma cells and cerebrocortical cultures". Arch. Biochem. Biophys. 331 (2): 208–214. doi:10.1006/abbi.1996.0300. PMID 8660700.
- Igarashi, Tomoji; Aritake, Shiro; Yasumoto, Takeshi (1999). "Mechanisms underlying the hemolytic and ichthyotoxic activities of maitotoxin". Natural Toxins. Wiley. 7 (2): 71–79. doi:10.1002/(sici)1522-7189(199903/04)7:2<71::aid-nt40>3.0.co;2-0. ISSN 1056-9014. PMID 10495469.
- Murata, M; et al. (1994). "Structure and partial stereochemical assignments for maitotoxin, the most toxic and largest natural non-biopolymer". J. Am. Chem. Soc. 116 (16): 7098–7107. doi:10.1021/ja00095a013.
- Sasaki, M; et al. (1996). "The complete structure of maitotoxin, I; Configuration of the C1-C14 side chain". Angew. Chem. Int. Ed. Engl. 35 (15): 1672–1675. doi:10.1002/anie.199616721.
- Kishi, Y (1998). "Complete structure of maitotoxin". Pure Appl. Chem. 70 (2): 339–344. doi:10.1351/pac199870020339.
- Gallimore AR, Spencer JB (2006). "Stereochemical Uniformity in Marine Polyether Ladders—Implications for the Biosynthesis and Structure of Maitotoxin". Angew. Chem. Int. Ed. Engl. 45 (27): 4406–4413. doi:10.1002/anie.200504284. PMID 16767782.
- Nicolaou KC, Frederick MO (2007). "On the structure of maitotoxin". Angew. Chem. Int. Ed. Engl. 46 (28): 5278–82. doi:10.1002/anie.200604656. PMID 17469088.
- Nicolaou K. C., Cole Kevin P., Frederick Michael O., Aversa Robert J., Denton Ross M. (2007). "Chemical Synthesis of the GHIJK Ring System and Further Experimental Support for the Originally Assigned Structure of Maitotoxin". Angew. Chem. Int. Ed. 46 (46): 8875–8879. doi:10.1002/anie.200703742. PMID 17943950.
{{cite journal}}: CS1 maint: multiple names: authors list (link) - Nicolaou K. C. (2008). "Chemical Synthesis of the GHIJKLMNO Ring System of Maitotoxin". Journal of the American Chemical Society. 130 (23): 7466–7476. doi:10.1021/ja801139f. PMID 18481856.
- Nicolaou K. C. (2010). "Synthesis of the ABCDEFG Ring System of Maitotoxin". Journal of the American Chemical Society. 132 (19): 6855–6861. doi:10.1021/ja102260q. PMC 2880607. PMID 20415445.
- Nicolaou K. C. (2014). "Synthesis and Biological Evaluation of QRSTUVWXYZA′ Domains of Maitotoxin". Journal of the American Chemical Society. 136 (46): 16444–16451. doi:10.1021/ja509829e. PMC 4244842. PMID 25374117.
- Chemistry's toughest total synthesis challenge put on hold by lack of funds Katrina Kramer 15 January 2015 Chemistry World http://www.rsc.org/chemistryworld/2015/01/chemistry-grandest-total-synthesis-challenge-maitotoxin-put-hold-lack-funds
Further reading
- JP JOSHI, Maitland (2004). Organic Chemistry, Third Edition. W. W. Norton & Company. ISBN 978-0-393-92408-4.