Cyanobacteria
Cyanobacteria (/saɪˌænoʊbækˈtɪəri.ə/), also known as Cyanophyta, are a phylum of gram-negative bacteria[4] that obtain energy via photosynthesis. The name cyanobacteria refers to their color (from Ancient Greek κυανός (kuanós) 'blue'),[5][6] which similarly forms the basis of cyanobacteria's common name, blue-green algae.[7][8][9][note 1] They appear to have originated in a freshwater or terrestrial environment.[10] Sericytochromatia, the proposed name of the paraphyletic and most basal group, is the ancestor of both the non-photosynthetic group Melainabacteria and the photosynthetic cyanobacteria, also called Oxyphotobacteria.[11]
| Cyanobacteria Temporal range: | |
|---|---|
 | |
| Microscope image of Cylindrospermum, a filamentous genus of cyanobacteria | |
| Scientific classification | |
| Domain: | Bacteria |
| (unranked): | Terrabacteria |
| (unranked): | Cyanobacteria-Melainabacteria group |
| Phylum: | Cyanobacteria Stanier, 1973 |
| Class: | Cyanophyceae |
| Orders[1] | |
|
As of 2014 the taxonomy was under revision[2][3]
| |
| Synonyms | |
|
List
| |
Cyanobacteria use photosynthetic pigments, such as carotenoids, phycobilins, and various forms of chlorophyll, which absorb energy from light. Unlike heterotrophic prokaryotes, cyanobacteria have internal membranes. These are flattened sacs called thylakoids where photosynthesis is performed.[12][13] Phototrophic eukaryotes such as green plants perform photosynthesis in plastids that are thought to have their ancestry in cyanobacteria, acquired long ago via a process called endosymbiosis. These endosymbiotic cyanobacteria in eukaryotes then evolved and differentiated into specialized organelles such as chloroplasts, chromoplasts, etioplasts, and leucoplasts, collectively known as plastids.
Cyanobacteria are the first organisms known to have produced oxygen. By producing and releasing oxygen as a byproduct of photosynthesis, cyanobacteria are thought to have converted the early oxygen-poor, reducing atmosphere into an oxidizing one, causing the Great Oxidation Event and the "rusting of the Earth",[14] which dramatically changed the composition of the Earth's life forms.[15]
The cyanobacteria Synechocystis and Cyanothece are important model organisms with potential applications in biotechnology for bioethanol production, food colorings, as a source of human and animal food, dietary supplements and raw materials.[16] Cyanobacteria produce a range of toxins known as cyanotoxins that can pose a danger to humans and animals.
Overview

Cyanobacteria are a very large and diverse phylum of photoautotrophic prokaryotes.[18] They are defined by their unique combination of pigments and their ability to perform oxygenic photosynthesis. They often live in colonial aggregates that can take on a multitude of forms.[19] Of particular interest are the filamentous species, which often dominate the upper layers of microbial mats found in extreme environments such as hot springs, hypersaline water, deserts and the polar regions,[20] but are also widely distributed in more mundane environments as well.[21]
Cyanobacteria are a group of photosynthetic bacteria evolutionarily optimized for environmental conditions of low oxygen.[22] Some species are nitrogen-fixing and live in a wide variety of moist soils and water, either freely or in a symbiotic relationship with plants or lichen-forming fungi (as in the lichen genus Peltigera).[23] They range from unicellular to filamentous and include colonial species. Colonies may form filaments, sheets, or even hollow spheres.
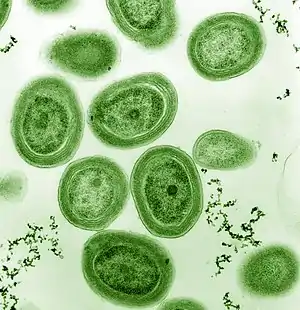
Cyanobacteria are globally widespread photosynthetic prokaryotes and are major contributors to global biogeochemical cycles.[24] They are the only oxygenic photosynthetic prokaryotes, and prosper in diverse and extreme habitats.[25] They are among the oldest organisms on Earth with fossil records dating back 3.5 billion years.[26] Since then, cyanobacteria have been essential players in the Earth's ecosystems. Planktonic cyanobacteria are a fundamental component of marine food webs and are major contributors to global carbon and nitrogen fluxes.[27][28] Some cyanobacteria form harmful algal blooms causing the disruption of aquatic ecosystem services and intoxication of wildlife and humans by the production of powerful toxins (cyanotoxins) such as microcystins, saxitoxin, and cylindrospermopsin.[29][30] Nowadays, cyanobacterial blooms pose a serious threat to aquatic environments and public health, and are increasing in frequency and magnitude globally.[31][24]
Cyanobacteria are ubiquitous in marine environments and play important roles as primary producers. Marine phytoplankton today contribute almost half of the Earth's total primary production.[32] Within the cyanobacteria, only a few lineages colonized the open-ocean (i.e., Crocosphaera and relatives, cyanobacterium UCYN-A, Trichodesmium, as well as Prochlorococcus and Synechococcus).[33][34][35][36] From these lineages, nitrogen fixing cyanobacteria are particularly important because they exert a control on primary productivity and the export of organic carbon to the deep ocean,[33] by converting nitrogen gas into ammonium, which is later used to make amino acids and proteins. Marine picocyanobacteria (i.e., Prochlorococcus and Synechococcus) numerically dominate most phytoplankton assemblages in modern oceans contributing importantly to primary productivity.[35][36][37] While some planktonic cyanobacteria are unicellular and free living cells (e.g., Crocosphaera, Prochlorococcus, Synechococcus), others have established symbiotic relationships with haptophyte algae, such as coccolithophores.[34] Amongst the filamentous forms, Trichodesmium are free-living and form aggregates. However, filamentous heterocyst-forming cyanobacteria (e.g., Richelia, Calothrix) are found in association with diatoms such as Hemiaulus, Rhizosolenia and Chaetoceros.[38][39][40][41]
Marine cyanobacteria include the smallest known photosynthetic organisms. The smallest of all, Prochlorococcus, is just 0.5 to 0.8 micrometres across.[42] In terms of individual numbers, Prochlorococcus is possibly the most plentiful species on Earth: a single millilitre of surface seawater can contain 100,000 cells or more. Worldwide there are estimated to be several octillion (1027) individuals.[43] Prochlorococcus is ubiquitous between 40°N and 40°S and dominates in the oligotrophic (nutrient poor) regions of the oceans.[44] The bacterium accounts for about 20% of the oxygen in the Earth's atmosphere.[45]
Morphology
Cyanobacteria are variable in morphology, ranging from unicellular and filamentous to colonial forms. Filamentous forms exhibit functional cell differentiation such as heterocysts (for nitrogen fixation), akinetes (resting stage cells), and hormogonia (reproductive, motile filaments). These, together with the intercellular connections they possess, are considered the first signs of multicellularity.[46][47][48][24]
Many cyanobacteria form motile filaments of cells, called hormogonia, that travel away from the main biomass to bud and form new colonies elsewhere.[49][50] The cells in a hormogonium are often thinner than in the vegetative state, and the cells on either end of the motile chain may be tapered. To break away from the parent colony, a hormogonium often must tear apart a weaker cell in a filament, called a necridium.
scale bars about 10 µm

• Non-heterocytous: (c) Arthrospira maxima,
Some filamentous species can differentiate into several different cell types:
- Vegetative cells – the normal, photosynthetic cells that are formed under favorable growing conditions
- Akinetes – climate-resistant spores that may form when environmental conditions become harsh
- Thick-walled heterocysts – which contain the enzyme nitrogenase vital for nitrogen fixation[52][53][54] in an anaerobic environment due to its sensitivity to oxygen.[54]
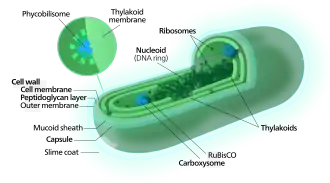
Each individual cell (each single cyanobacterium) typically has a thick, gelatinous cell wall.[55] They lack flagella, but hormogonia of some species can move about by gliding along surfaces.[56] Many of the multicellular filamentous forms of Oscillatoria are capable of a waving motion; the filament oscillates back and forth. In water columns, some cyanobacteria float by forming gas vesicles, as in archaea.[57] These vesicles are not organelles as such. They are not bounded by lipid membranes, but by a protein sheath.
Nitrogen fixation
Some cyanobacteria can fix atmospheric nitrogen in anaerobic conditions by means of specialized cells called heterocysts.[53][54] Heterocysts may also form under the appropriate environmental conditions (anoxic) when fixed nitrogen is scarce. Heterocyst-forming species are specialized for nitrogen fixation and are able to fix nitrogen gas into ammonia (NH3), nitrites (NO−2) or nitrates (NO−3), which can be absorbed by plants and converted to protein and nucleic acids (atmospheric nitrogen is not bioavailable to plants, except for those having endosymbiotic nitrogen-fixing bacteria, especially the family Fabaceae, among others).
Free-living cyanobacteria are present in the water of rice paddies, and cyanobacteria can be found growing as epiphytes on the surfaces of the green alga, Chara, where they may fix nitrogen.[58] Cyanobacteria such as Anabaena (a symbiont of the aquatic fern Azolla) can provide rice plantations with biofertilizer.[59]
Photosynthesis
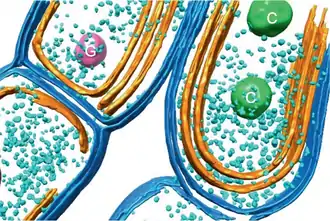
Carbon fixation
Cyanobacteria use the energy of sunlight to drive photosynthesis, a process where the energy of light is used to synthesize organic compounds from carbon dioxide. Because they are aquatic organisms, they typically employ several strategies which are collectively known as a "CO2 concentrating mechanism" to aid in the acquisition of inorganic carbon (CO2 or bicarbonate). Among the more specific strategies is the widespread prevalence of the bacterial microcompartments known as carboxysomes,[61] which co-operate with active transporters of CO2 and bicarbonate, in order to accumulate bicarbonate into the cytoplasm of the cell.[62] Carboxysomes are icosahedral structures composed of hexameric shell proteins that assemble into cage-like structures that can be several hundreds of nanometres in diameter. It is believed that these structures tether the CO2-fixing enzyme, RuBisCO, to the interior of the shell, as well as the enzyme carbonic anhydrase, using metabolic channeling to enhance the local CO2 concentrations and thus increase the efficiency of the RuBisCO enzyme.[63]
Electron transport
In contrast to purple bacteria and other bacteria performing anoxygenic photosynthesis, thylakoid membranes of cyanobacteria are not continuous with the plasma membrane but are separate compartments.[64] The photosynthetic machinery is embedded in the thylakoid membranes, with phycobilisomes acting as light-harvesting antennae attached to the membrane, giving the green pigmentation observed (with wavelengths from 450 nm to 660 nm) in most cyanobacteria.[65]
While most of the high-energy electrons derived from water are used by the cyanobacterial cells for their own needs, a fraction of these electrons may be donated to the external environment via electrogenic activity.[66]
Respiration
Respiration in cyanobacteria can occur in the thylakoid membrane alongside photosynthesis,[67] with their photosynthetic electron transport sharing the same compartment as the components of respiratory electron transport. While the goal of photosynthesis is to store energy by building carbohydrates from CO2, respiration is the reverse of this, with carbohydrates turned back into CO2 accompanying energy release.
Cyanobacteria appear to separate these two processes with their plasma membrane containing only components of the respiratory chain, while the thylakoid membrane hosts an interlinked respiratory and photosynthetic electron transport chain.[67] Cyanobacteria use electrons from succinate dehydrogenase rather than from NADPH for respiration.[67]
Cyanobacteria only respire during the night (or in the dark) because the facilities used for electron transport are used in reverse for photosynthesis while in the light.[68]
Electron transport chain
Many cyanobacteria are able to reduce nitrogen and carbon dioxide under aerobic conditions, a fact that may be responsible for their evolutionary and ecological success. The water-oxidizing photosynthesis is accomplished by coupling the activity of photosystem (PS) II and I (Z-scheme). In contrast to green sulfur bacteria which only use one photosystem, the use of water as an electron donor is energetically demanding, requiring two photosystems.[69]
Attached to the thylakoid membrane, phycobilisomes act as light-harvesting antennae for the photosystems.[70] The phycobilisome components (phycobiliproteins) are responsible for the blue-green pigmentation of most cyanobacteria.[71] The variations on this theme are due mainly to carotenoids and phycoerythrins that give the cells their red-brownish coloration. In some cyanobacteria, the color of light influences the composition of the phycobilisomes.[72][73] In green light, the cells accumulate more phycoerythrin, which absorbs green light, whereas in red light they produce more phycocyanin which absorbs red. Thus, these bacteria can change from brick-red to bright blue-green depending on whether they are exposed to green light or to red light.[74] This process of "complementary chromatic adaptation" is a way for the cells to maximize the use of available light for photosynthesis.
A few genera lack phycobilisomes and have chlorophyll b instead (Prochloron, Prochlorococcus, Prochlorothrix). These were originally grouped together as the prochlorophytes or chloroxybacteria, but appear to have developed in several different lines of cyanobacteria. For this reason, they are now considered as part of the cyanobacterial group.[75][76]
Metabolism
In general, photosynthesis in cyanobacteria uses water as an electron donor and produces oxygen as a byproduct, though some may also use hydrogen sulfide[77] a process which occurs among other photosynthetic bacteria such as the purple sulfur bacteria.
Carbon dioxide is reduced to form carbohydrates via the Calvin cycle.[78] The large amounts of oxygen in the atmosphere are considered to have been first created by the activities of ancient cyanobacteria.[79] They are often found as symbionts with a number of other groups of organisms such as fungi (lichens), corals, pteridophytes (Azolla), angiosperms (Gunnera), etc.[80]
There are some groups capable of heterotrophic growth,[81] while others are parasitic, causing diseases in invertebrates or algae (e.g., the black band disease).[82][83][84]
Ecology
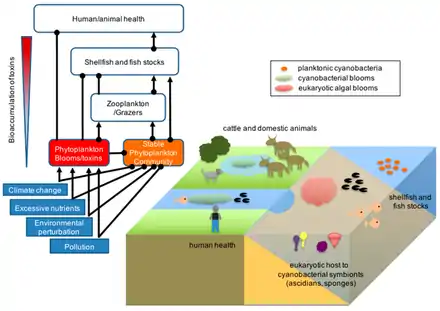
Cyanobacteria can be found in almost every terrestrial and aquatic habitat – oceans, fresh water, damp soil, temporarily moistened rocks in deserts, bare rock and soil, and even Antarctic rocks. They can occur as planktonic cells or form phototrophic biofilms. They are found inside stones and shells (in endolithic ecosystems).[86] A few are endosymbionts in lichens, plants, various protists, or sponges and provide energy for the host. Some live in the fur of sloths, providing a form of camouflage.[87]
Aquatic cyanobacteria are known for their extensive and highly visible blooms that can form in both freshwater and marine environments. The blooms can have the appearance of blue-green paint or scum. These blooms can be toxic, and frequently lead to the closure of recreational waters when spotted. Marine bacteriophages are significant parasites of unicellular marine cyanobacteria.[88]
Cyanobacterial growth is favoured in ponds and lakes where waters are calm and have little turbulent mixing.[89] Their lifecycles are disrupted when the water naturally or artificially mixes from churning currents caused by the flowing water of streams or the churning water of fountains. For this reason blooms of cyanobacteria seldom occur in rivers unless the water is flowing slowly. Growth is also favoured at higher temperatures which enable Microcystis species to outcompete diatoms and green algae, and potentially allow development of toxins.[89]
Based on environmental trends, models and observations suggest cyanobacteria will likely increase their dominance in aquatic environments. This can lead to serious consequences, particularly the contamination of sources of drinking water. Researchers including Linda Lawton at Robert Gordon University, have developed techniques to study these.[90] Cyanobacteria can interfere with water treatment in various ways, primarily by plugging filters (often large beds of sand and similar media) and by producing cyanotoxins, which have the potential to cause serious illness if consumed. Consequences may also lie within fisheries and waste management practices. Anthropogenic eutrophication, rising temperatures, vertical stratification and increased atmospheric carbon dioxide are contributors to cyanobacteria increasing dominance of aquatic ecosystems.[91]
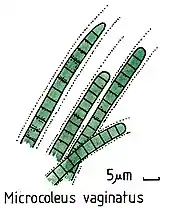
Cyanobacteria have been found to play an important role in terrestrial habitats. It has been widely reported that cyanobacteria soil crusts help to stabilize soil to prevent erosion and retain water.[92] An example of a cyanobacterial species that does so is Microcoleus vaginatus. M. vaginatus stabilizes soil using a polysaccharide sheath that binds to sand particles and absorbs water.[93]
Some of these organisms contribute significantly to global ecology and the oxygen cycle. The tiny marine cyanobacterium Prochlorococcus was discovered in 1986 and accounts for more than half of the photosynthesis of the open ocean.[94] Circadian rhythms were once thought to only exist in eukaryotic cells but many cyanobacteria display a bacterial circadian rhythm.
"Cyanobacteria are arguably the most successful group of microorganisms on earth. They are the most genetically diverse; they occupy a broad range of habitats across all latitudes, widespread in freshwater, marine, and terrestrial ecosystems, and they are found in the most extreme niches such as hot springs, salt works, and hypersaline bays. Photoautotrophic, oxygen-producing cyanobacteria created the conditions in the planet's early atmosphere that directed the evolution of aerobic metabolism and eukaryotic photosynthesis. Cyanobacteria fulfill vital ecological functions in the world's oceans, being important contributors to global carbon and nitrogen budgets." – Stewart and Falconer[95]
Cyanobionts
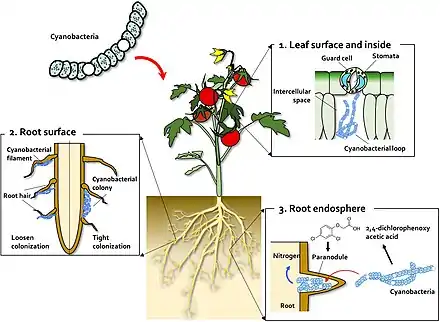
(2) On the root surface, cyanobacteria exhibit two types of colonization pattern; in the root hair, filaments of Anabaena and Nostoc species form loose colonies, and in the restricted zone on the root surface, specific Nostoc species form cyanobacterial colonies.
(3) Co-inoculation with 2,4-D and Nostoc spp. increases para-nodule formation and nitrogen fixation. A large number of Nostoc spp. isolates colonize the root endosphere and form para-nodules.[96]
Some cyanobacteria, the so-called cyanobionts (cyanobacterial symbionts), have a symbiotic relationship with other organisms, both unicellular and multicellular.[97] As illustrated on the right, there are many examples of cyanobacteria interacting symbiotically with land plants.[98][99][100][101] Cyanobacteria can enter the plant through the stomata and colonize the intercellular space, forming loops and intracellular coils.[102] Anabaena spp. colonize the roots of wheat and cotton plants.[103][104][105] Calothrix sp. has also been found on the root system of wheat.[104][105] Monocots, such as wheat and rice, have been colonised by Nostoc spp.,[106][107][108][109] In 1991, Ganther and others isolated diverse heterocystous nitrogen-fixing cyanobacteria, including Nostoc, Anabaena and Cylindrospermum, from plant root and soil. Assessment of wheat seedling roots revealed two types of association patterns: loose colonization of root hair by Anabaena and tight colonization of the root surface within a restricted zone by Nostoc.[106][96]
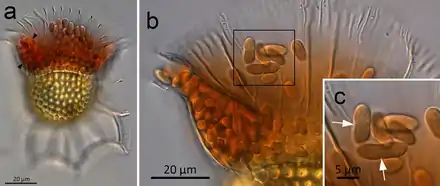
(a) O. magnificus with numerous cyanobionts present in the upper and lower girdle lists (black arrowheads) of the cingulum termed the symbiotic chamber.
(b) O. steinii with numerous cyanobionts inhabiting the symbiotic chamber.
(c) Enlargement of the area in (b) showing two cyanobionts that are being divided by binary transverse fission (white arrows).

The relationships between cyanobionts (cyanobacterial symbionts) and protistan hosts are particularly noteworthy, as some nitrogen-fixing cyanobacteria (diazotrophs) play an important role in primary production, especially in nitrogen-limited oligotrophic oceans.[110][111][112] Cyanobacteria, mostly pico-sized Synechococcus and Prochlorococcus, are ubiquitously distributed and are the most abundant photosynthetic organisms on Earth, accounting for a quarter of all carbon fixed in marine ecosystems.[37][113][114] In contrast to free-living marine cyanobacteria, some cyanobionts are known to be responsible for nitrogen fixation rather than carbon fixation in the host.[115][116] However, the physiological functions of most cyanobionts remain unknown. Cyanobionts have been found in numerous protist groups, including dinoflagellates, tintinnids, radiolarians, amoebae, diatoms, and haptophytes.[117][118] Among these cyanobionts, little is known regarding the nature (e.g., genetic diversity, host or cyanobiont specificity, and cyanobiont seasonality) of the symbiosis involved, particularly in relation to dinoflagellate host.[97]
Collective behaviour
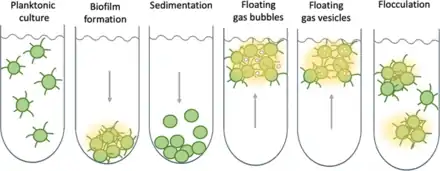
Some cyanobacteria – even single-celled ones – show striking collective behaviours and form colonies (or blooms) that can float on water and have important ecological roles. For instance, billions of years ago, communities of marine Paleoproterozoic cyanobacteria could have helped create the biosphere as we know it by burying carbon compounds and allowing the initial build-up of oxygen in the atmosphere.[120] On the other hand, toxic cyanobacterial blooms are an increasing issue for society, as their toxins can be harmful to animals.[31] Extreme blooms can also deplete water of oxygen and reduce the penetration of sunlight and visibility, thereby compromising the feeding and mating behaviour of light-reliant species.[119]
As shown in the diagram on the right, bacteria can stay in suspension as individual cells, adhere collectively to surfaces to form biofilms, passively sediment, or flocculate to form suspended aggregates. Cyanobacteria are able to produce sulphated polysaccharides (yellow haze surrounding clumps of cells) that enable them to form floating aggregates. In 2021, Maeda et al. discovered that oxygen produced by cyanobacteria becomes trapped in the network of polysaccharides and cells, enabling the microorganisms to form buoyant blooms.[121] It is thought that specific protein fibres known as pili (represented as lines radiating from the cells) may act as an additional way to link cells to each other or onto surfaces. Some cyanobacteria also use sophisticated intracellular gas vesicles as floatation aids.[119]
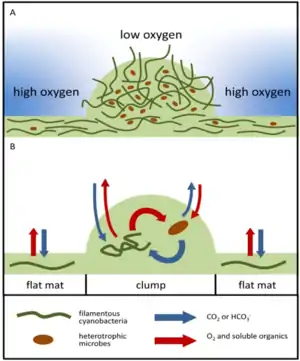
The diagram on the left above shows a proposed model of microbial distribution, spatial organization, carbon and O2 cycling in clumps and adjacent areas. (a) Clumps contain denser cyanobacterial filaments and heterotrophic microbes. The initial differences in density depend on cyanobacterial motility and can be established over short timescales. Darker blue color outside of the clump indicates higher oxygen concentrations in areas adjacent to clumps. Oxic media increase the reversal frequencies of any filaments that begin to leave the clumps, thereby reducing the net migration away from the clump. This enables the persistence of the initial clumps over short timescales; (b) Spatial coupling between photosynthesis and respiration in clumps. Oxygen produced by cyanobacteria diffuses into the overlying medium or is used for aerobic respiration. Dissolved inorganic carbon (DIC) diffuses into the clump from the overlying medium and is also produced within the clump by respiration. In oxic solutions, high O2 concentrations reduce the efficiency of CO2 fixation and result in the excretion of glycolate. Under these conditions, clumping can be beneficial to cyanobacteria if it stimulates the retention of carbon and the assimilation of inorganic carbon by cyanobacteria within clumps. This effect appears to promote the accumulation of particulate organic carbon (cells, sheaths and heterotrophic organisms) in clumps.[122]
It has been unclear why and how cyanobacteria form communities. Aggregation must divert resources away from the core business of making more cyanobacteria, as it generally involves the production of copious quantities of extracellular material. In addition, cells in the centre of dense aggregates can also suffer from both shading and shortage of nutrients.[123][124] So, what advantage does this communal life bring for cyanobacteria?[119]
New insights into how cyanobacteria form blooms have come from a 2021 study on the cyanobacterium Synechocystis. These use a set of genes that regulate the production and export of sulphated polysaccharides, chains of sugar molecules modified with sulphate groups that can often be found in marine algae and animal tissue. Many bacteria generate extracellular polysaccharides, but sulphated ones have only been seen in cyanobacteria. In Synechocystis these sulphated polysaccharide help the cyanobacterium form buoyant aggregates by trapping oxygen bubbles in the slimy web of cells and polysaccharides.[121][119]
Previous studies on Synechocystis have shown type IV pili, which decorate the surface of cyanobacteria, also play a role in forming blooms.[126][123] These retractable and adhesive protein fibres are important for motility, adhesion to substrates and DNA uptake.[127] The formation of blooms may require both type IV pili and Synechan – for example, the pili may help to export the polysaccharide outside the cell. Indeed, the activity of these protein fibres may be connected to the production of extracellular polysaccharides in filamentous cyanobacteria.[128] A more obvious answer would be that pili help to build the aggregates by binding the cells with each other or with the extracellular polysaccharide. As with other kinds of bacteria,[129] certain components of the pili may allow cyanobacteria from the same species to recognise each other and make initial contacts, which are then stabilised by building a mass of extracellular polysaccharide.[119]
The bubble flotation mechanism identified by Maeda et al. joins a range of known strategies that enable cyanobacteria to control their buoyancy, such as using gas vesicles or accumulating carbohydrate ballasts.[130] Type IV pili on their own could also control the position of marine cyanobacteria in the water column by regulating viscous drag.[131] Extracellular polysaccharide appears to be a multipurpose asset for cyanobacteria, from floatation device to food storage, defence mechanism and mobility aid.[128][119]
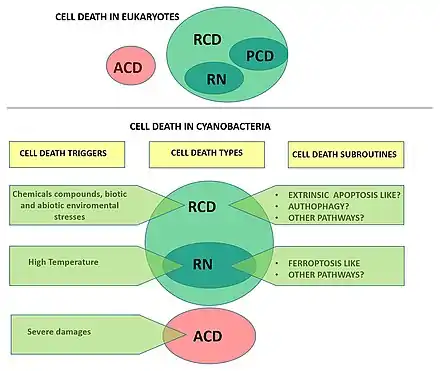
Cellular death
One of the most critical processes determining cyanobacterial eco-physiology is cellular death. Evidence supports the existence of controlled cellular demise in cyanobacteria, and various forms of cell death have been described as a response to biotic and abiotic stresses. However, cell death research in cyanobacteria is a relatively young field and understanding of the underlying mechanisms and molecular machinery underpinning this fundamental process remains largely elusive.[24] However, reports on cell death of marine and freshwater cyanobacteria indicate this process has major implications for the ecology of microbial communities/[132][133][134][135] Different forms of cell demise have been observed in cyanobacteria under several stressful conditions,[136][137] and cell death has been suggested to play a key role in developmental processes, such as akinete and heterocyst differentiation.[138][46][24]
Cyanophages
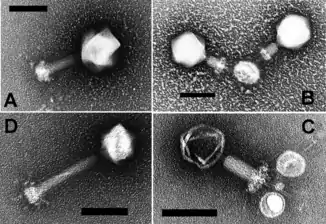
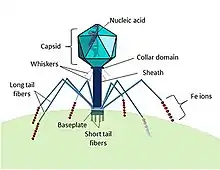
Cyanophages are viruses that infect cyanobacteria. Cyanophages can be found in both freshwater and marine environments.[139] Marine and freshwater cyanophages have icosahedral heads, which contain double-stranded DNA, attached to a tail by connector proteins.[140] The size of the head and tail vary among species of cyanophages. Cyanophages like other bacteriophages rely on Brownian motion to collide with bacteria, and then use receptor binding proteins to recognize cell surface proteins, which leads to adherence. Viruses with contractile tails then rely on receptors found on their tails to recognize highly conserved proteins on the surface of the host cell.[141]
Cyanophages infect a wide range of cyanobacteria and are key regulators of the cyanobacterial populations in aquatic environments, and may aid in the prevention of cyanobacterial blooms in freshwater and marine ecosystems. These blooms can pose a danger to humans and other animals, particularly in eutrophic freshwater lakes. Infection by these viruses is highly prevalent in cells belonging to Synechococcus spp. in marine environments, where up to 5% of cells belonging to marine cyanobacterial cells have been reported to contain mature phage particles.[142]
The first cyanophage, LPP-1, was discovered in 1963.[143] Cyanophages are classified within the bacteriophage families Myoviridae (e.g. AS-1, N-1), Podoviridae (e.g. LPP-1) and Siphoviridae (e.g. S-1).[143]
Movement
It has long been known that filamentous cyanobacteria perform surface motions, and that these movements result from type IV pili.[144][128][145] Additionally, Synechococcus, a marine cyanobacteria, is known to swim at a speed of 25 μm/s by a mechanism different to that of bacterial flagella.[146] Formation of waves on the cyanobacteria surface is thought to push surrounding water backwards.[147][148] Cells are known to be motile by a gliding method[149] and a novel uncharacterized, nonphototactic swimming method[150] that does not involve flagellar motion.
Many species of cyanobacteria are capable of gliding. Gliding is a form of cell movement that differs from crawling or swimming in that it does not rely on any obvious external organ or change in cell shape and it occurs only in the presence of a substrate.[151][152] Gliding in filamentous cyanobacteria appears to be powered by a "slime jet" mechanism, in which the cells extrude a gel that expands quickly as it hydrates providing a propulsion force,[153][154] although some unicellular cyanobacteria use type IV pili for gliding.[155][21]
Cyanobacteria have strict light requirements. Too little light can result in insufficient energy production, and in some species may cause the cells to resort to heterotrophic respiration.[20] Too much light can inhibit the cells, decrease photosynthesis efficiency and cause damage by bleaching. UV radiation is especially deadly for cyanobacteria, with normal solar levels being significantly detrimental for these microorganisms in some cases.[19][156][21]
Filamentous cyanobacteria that live in microbial mats often migrate vertically and horizontally within the mat in order to find an optimal niche that balances their light requirements for photosynthesis against their sensitivity to photodamage. For example, the filamentous cyanobacteria Oscillatoria sp. and Spirulina subsalsa found in the hypersaline benthic mats of Guerrero Negro, Mexico migrate downwards into the lower layers during the day in order to escape the intense sunlight and then rise to the surface at dusk.[157] In contrast, the population of Microcoleus chthonoplastes found in hypersaline mats in Camargue, France migrate to the upper layer of the mat during the day and are spread homogenously through the mat at night.[158] An in vitro experiment using P. uncinatum also demonstrated this species' tendency to migrate in order to avoid damaging radiation.[19][156] These migrations are usually the result of some sort of photomovement, although other forms of taxis can also play a role.[159][21]
Photomovement – the modulation of cell movement as a function of the incident light – is employed by the cyanoabacteria as a means to find optimal light conditions in their environment. There are three types of photomovement: photokinesis, phototaxis and photophobic responses.[160][161][162][21]
Photokinetic microorganisms modulate their gliding speed according to the incident light intensity. For example, the speed with which Phormidium autumnale glides increases linearly with the incident light intensity.[163][21]
Phototactic microorganisms move according to the direction of the light within the environment, such that positively phototactic species will tend to move roughly parallel to the light and towards the light source. Species such as Phormidium uncinatum cannot steer directly towards the light, but rely on random collisions to orient themselves in the right direction, after which they tend to move more towards the light source. Others, such as Anabaena variabilis, can steer by bending the trichome.[164][21]
Finally, photophobic microorganisms respond to spatial and temporal light gradients. A step-up photophobic reaction occurs when an organism enters a brighter area field from a darker one and then reverses direction, thus avoiding the bright light. The opposite reaction, called a step-down reaction, occurs when an organism enters a dark area from a bright area and then reverses direction, thus remaining in the light.[21]
Evolution
Earth history
Life timeline | ||||||||||||||||||||||||||||||||||||||||||||||
−4500 — – — – −4000 — – — – −3500 — – — – −3000 — – — – −2500 — – — – −2000 — – — – −1500 — – — – −1000 — – — – −500 — – — – 0 — | Water Photosynthesis Eukaryotes Multicellular life P l a n t s Arthropods Molluscs Flowers Dinosaurs Mammals Birds Primates H a d e a n A r c h e a n P r o t e r o z o i c P h a n e r o z o i c |
| ||||||||||||||||||||||||||||||||||||||||||||
(million years ago) *Ice Ages | ||||||||||||||||||||||||||||||||||||||||||||||
Stromatolites are layered biochemical accretionary structures formed in shallow water by the trapping, binding, and cementation of sedimentary grains by biofilms (microbial mats) of microorganisms, especially cyanobacteria.[165]
During the Precambrian, stromatolite communities of microorganisms grew in most marine and non-marine environments in the photic zone. After the Cambrian explosion of marine animals, grazing on the stromatolite mats by herbivores greatly reduced the occurrence of the stromatolites in marine environments. Since then, they are found mostly in hypersaline conditions where grazing invertebrates cannot live (e.g. Shark Bay, Western Australia). Stromatolites provide ancient records of life on Earth by fossil remains which date from 3.5 Ga ago.[166] As of 2010 the oldest undisputed evidence of cyanobacteria is from 2.1 Ga ago, but there is some evidence for them as far back as 2.7 Ga ago. Oxygen concentrations in the atmosphere remained around or below 1% of today's level until 2.4 Ga ago (the Great Oxygenation Event). The rise in oxygen may have caused a fall in the concentration of atmospheric methane, and triggered the Huronian glaciation from around 2.4 to 2.1 Ga ago. In this way, cyanobacteria may have killed off much of the other bacteria of the time.[167]
Oncolites are sedimentary structures composed of oncoids, which are layered structures formed by cyanobacterial growth. Oncolites are similar to stromatolites, but instead of forming columns, they form approximately spherical structures that were not attached to the underlying substrate as they formed.[168] The oncoids often form around a central nucleus, such as a shell fragment,[169] and a calcium carbonate structure is deposited by encrusting microbes. Oncolites are indicators of warm waters in the photic zone, but are also known in contemporary freshwater environments.[170] These structures rarely exceed 10 cm in diameter.
One former classification scheme of cyanobacterial fossils divided them into the porostromata and the spongiostromata. These are now recognized as form taxa and considered taxonomically obsolete; however, some authors have advocated for the terms remaining informally to describe form and structure of bacterial fossils.[171]
 Stromatolites left behind by cyanobacteria are the oldest known fossils of life on Earth. This fossil is one billion years old.
Stromatolites left behind by cyanobacteria are the oldest known fossils of life on Earth. This fossil is one billion years old._3.jpg.webp) Oncolitic limestone formed from successive layers of calcium carbonate precipitated by cyanobacteria
Oncolitic limestone formed from successive layers of calcium carbonate precipitated by cyanobacteria Oncolites from the Late Devonian Alamo bolide impact in Nevada
Oncolites from the Late Devonian Alamo bolide impact in Nevada Cyanobacterial remains of an annulated tubular microfossil Oscillatoriopsis longa [172]
Cyanobacterial remains of an annulated tubular microfossil Oscillatoriopsis longa [172]
Scale bar: 100 μm
Origin of photosynthesis
As far as we can tell, oxygenic photosynthesis only evolved once (in prokaryotic cyanobacteria), and all photosynthetic eukaryotes (including all plants and algae) have acquired this ability from them. In other words, all the oxygen that makes the atmosphere breathable for aerobic organisms originally comes from cyanobacteria or their later descendants.[173]
Cyanobacteria remained principal primary producers throughout the Proterozoic Eon (2500–543 Ma), in part because the redox structure of the oceans favored photoautotrophs capable of nitrogen fixation. Green algae joined blue-greens as major primary producers on continental shelves near the end of the Proterozoic, but only with the Mesozoic (251–65 Ma) radiations of dinoflagellates, coccolithophorids, and diatoms did primary production in marine shelf waters take modern form. Cyanobacteria remain critical to marine ecosystems as primary producers in oceanic gyres, as agents of biological nitrogen fixation, and, in modified form, as the plastids of marine eukaryotic algae.[174]
Origin of chloroplasts
Primary chloroplasts are cell organelles found in some eukaryotic lineages, where they are specialized in performing photosynthesis. They are considered to have evolved from endosymbiotic cyanobacteria.[175][176] After some years of debate,[177] it is now generally accepted that the three major groups of primary endosymbiotic eukaryotes (i.e. green plants, red algae and glaucophytes) form one large monophyletic group called Archaeplastida, which evolved after one unique endosymbiotic event.[178][179][180][181]
The morphological similarity between chloroplasts and cyanobacteria was first reported by German botanist Andreas Franz Wilhelm Schimper in the 19th century[182] Chloroplasts are only found in plants and algae,[183] thus paving the way for Russian biologist Konstantin Mereschkowski to suggest in 1905 the symbiogenic origin of the plastid.[184] Lynn Margulis brought this hypothesis back to attention more than 60 years later[185] but the idea did not become fully accepted until supplementary data started to accumulate. The cyanobacterial origin of plastids is now supported by various pieces of phylogenetic,[186][178][181] genomic,[187] biochemical[188][189] and structural evidence.[190] The description of another independent and more recent primary endosymbiosis event between a cyanobacterium and a separate eukaryote lineage (the rhizarian Paulinella chromatophora) also gives credibility to the endosymbiotic origin of the plastids.[191]
In addition to this primary endosymbiosis, many eukaryotic lineages have been subject to secondary or even tertiary endosymbiotic events, that is the "Matryoshka-like" engulfment by a eukaryote of another plastid-bearing eukaryote.[193][175]
Chloroplasts have many similarities with cyanobacteria, including a circular chromosome, prokaryotic-type ribosomes, and similar proteins in the photosynthetic reaction center.[194][195] The endosymbiotic theory suggests that photosynthetic bacteria were acquired (by endocytosis) by early eukaryotic cells to form the first plant cells. Therefore, chloroplasts may be photosynthetic bacteria that adapted to life inside plant cells. Like mitochondria, chloroplasts still possess their own DNA, separate from the nuclear DNA of their plant host cells and the genes in this chloroplast DNA resemble those in cyanobacteria.[196] DNA in chloroplasts codes for redox proteins such as photosynthetic reaction centers. The CoRR hypothesis proposes this co-location is required for redox regulation.
Marine origins
| Part of a series on |
| Plankton |
|---|
 |
|
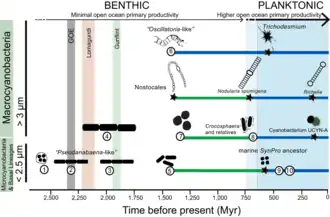
Cyanobacteria have fundamentally transformed the geochemistry of the planet.[200][197] Multiple lines of geochemical evidence support the occurrence of intervals of profound global environmental change at the beginning and end of the Proterozoic (2,500–542 Mya). [202][203] While it is widely accepted that the presence of molecular oxygen in the early fossil record was the result of cyanobacteria activity, little is known about how cyanobacteria evolution (e.g., habitat preference) may have contributed to changes in biogeochemical cycles through Earth history. Geochemical evidence has indicated that there was a first step-increase in the oxygenation of the Earth's surface, which is known as the Great Oxidation Event (GOE), in the early Paleoproterozoic (2,500–1,600 Mya).[200][197] A second but much steeper increase in oxygen levels, known as the Neoproterozoic Oxygenation Event (NOE),[202][204][205] occurred at around 800 to 500 Mya.[203][206] Recent chromium isotope data point to low levels of atmospheric oxygen in the Earth's surface during the mid-Proterozoic, which is consistent with the late evolution of marine planktonic cyanobacteria during the Cryogenian;[207] both types of evidence help explain the late emergence and diversification of animals.[208][41]
Understanding the evolution of planktonic cyanobacteria is important because their origin fundamentally transformed the nitrogen and carbon cycles towards the end of the Pre-Cambrian.[206] It remains unclear, however, what evolutionary events led to the emergence of open-ocean planktonic forms within cyanobacteria and how these events relate to geochemical evidence during the Pre-Cambrian.[202] So far, it seems that ocean geochemistry (e.g., euxinic conditions during the early- to mid-Proterozoic)[202][205][209] and nutrient availability [210] likely contributed to the apparent delay in diversification and widespread colonization of open ocean environments by planktonic cyanobacteria during the Neoproterozoic.[206][41]
Genetics
Cyanobacteria are capable of natural genetic transformation.[211][212][213] Natural genetic transformation is the genetic alteration of a cell resulting from the direct uptake and incorporation of exogenous DNA from its surroundings. For bacterial transformation to take place, the recipient bacteria must be in a state of competence, which may occur in nature as a response to conditions such as starvation, high cell density or exposure to DNA damaging agents. In chromosomal transformation, homologous transforming DNA can be integrated into the recipient genome by homologous recombination, and this process appears to be an adaptation for repairing DNA damage.[214]
DNA repair
Cyanobacteria are challenged by environmental stresses and internally generated reactive oxygen species that cause DNA damage. Cyanobacteria possess numerous E. coli-like DNA repair genes.[215] Several DNA repair genes are highly conserved in cyanobacteria, even in small genomes, suggesting that core DNA repair processes such as recombinational repair, nucleotide excision repair and methyl-directed DNA mismatch repair are common among cyanobacteria.[215]
Classification
Phylogeny
| 16S rRNA based LTP_12_2021[216][217][218] | GTDB 07-RS207 by Genome Taxonomy Database[219][220][221] | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
Taxonomy

Historically, bacteria were first classified as plants constituting the class Schizomycetes, which along with the Schizophyceae (blue-green algae/Cyanobacteria) formed the phylum Schizophyta,[222] then in the phylum Monera in the kingdom Protista by Haeckel in 1866, comprising Protogens, Protamaeba, Vampyrella, Protomonae, and Vibrio, but not Nostoc and other cyanobacteria, which were classified with algae,[223] later reclassified as the Prokaryotes by Chatton.[224]
The cyanobacteria were traditionally classified by morphology into five sections, referred to by the numerals I–V. The first three – Chroococcales, Pleurocapsales, and Oscillatoriales – are not supported by phylogenetic studies. The latter two – Nostocales and Stigonematales – are monophyletic, and make up the heterocystous cyanobacteria.[225][226]
The members of Chroococales are unicellular and usually aggregate in colonies. The classic taxonomic criterion has been the cell morphology and the plane of cell division. In Pleurocapsales, the cells have the ability to form internal spores (baeocytes). The rest of the sections include filamentous species. In Oscillatoriales, the cells are uniseriately arranged and do not form specialized cells (akinetes and heterocysts).[227] In Nostocales and Stigonematales, the cells have the ability to develop heterocysts in certain conditions. Stigonematales, unlike Nostocales, include species with truly branched trichomes.[225]
Most taxa included in the phylum or division Cyanobacteria have not yet been validly published under The International Code of Nomenclature of Prokaryotes (ICNP) except:
- The classes Chroobacteria, Hormogoneae, and Gloeobacteria
- The orders Chroococcales, Gloeobacterales, Nostocales, Oscillatoriales, Pleurocapsales, and Stigonematales
- The families Prochloraceae and Prochlorotrichaceae
- The genera Halospirulina, Planktothricoides, Prochlorococcus, Prochloron, and Prochlorothrix
The remainder are validly published under the International Code of Nomenclature for algae, fungi, and plants.
Formerly, some bacteria, like Beggiatoa, were thought to be colorless Cyanobacteria.[228]
The currently accepted taxonomy is based on the List of Prokaryotic names with Standing in Nomenclature (LPSN)[229] and National Center for Biotechnology Information (NCBI).[230] Class "Cyanobacteriia"
- Subclass Gloeobacteria
- Gloeobacterales Cavalier-Smith 2002
- Subclass Phycobacteria
- "Elainellales"
- "Eurycoccales"
- Gloeoemargaritales Moreira et al. 2016
- "Leptolyngbyales"
- "Neosynechococcales"
- "Phormidesmiales"
- Prochlorococcaceae Komárek & Strunecky 2020 {"PCC-6307"}
- Pseudanabaenales Hoffmann, Komárek & Kastovsky 2005
- Thermostichales Komárek & Strunecký 2020
- "Thermosynechococcales"
- Nostocophycidae
- Cyanobacteriales Rippka & Cohen-Bazire 1983 (Chamaesiphonales, Chroococcales, Chroococcidiopsidales, Nostocales, Oscillatoriales, Pleurocapsales, Spirulinales, Stigonematales)
- Synechococcophycidae
- "Limnotrichales"
- Prochlorotrichaceae Burger-Wiersma et al. 1989 {PCC-9006}
- Synechococcales Hoffmann, Komárek & Kastovsky 2005
Relation to humans
Biotechnology

The unicellular cyanobacterium Synechocystis sp. PCC6803 was the third prokaryote and first photosynthetic organism whose genome was completely sequenced.[231] It continues to be an important model organism.[232] Cyanothece ATCC 51142 is an important diazotrophic model organism. The smallest genomes have been found in Prochlorococcus spp. (1.7 Mb)[233][234] and the largest in Nostoc punctiforme (9 Mb).[235] Those of Calothrix spp. are estimated at 12–15 Mb,[236] as large as yeast.
Recent research has suggested the potential application of cyanobacteria to the generation of renewable energy by directly converting sunlight into electricity. Internal photosynthetic pathways can be coupled to chemical mediators that transfer electrons to external electrodes.[237][238] In the shorter term, efforts are underway to commercialize algae-based fuels such as diesel, gasoline, and jet fuel.[66][239][240] Cyanobacteria have been also engineered to produce ethanol[241] and experiments have shown that when one or two CBB genes are being over expressed, the yield can be even higher.[242][243]
Cyanobacteria may possess the ability to produce substances that could one day serve as anti-inflammatory agents and combat bacterial infections in humans.[244] Cyanobacteria's photosynthetic output of sugar and oxygen has been demonstrated to have therapeutic value in rats with heart attacks.[245] While cyanobacteria can naturally produce various secondary metabolites, they can serve as advantageous hosts for plant-derived metabolites production owing to biotechnological advances in systems biology and synthetic biology.[246]
Spirulina's extracted blue color is used as a natural food coloring.[247]
Researchers from several space agencies argue that cyanobacteria could be used for producing goods for human consumption in future crewed outposts on Mars, by transforming materials available on this planet.[248]
Human nutrition

Some cyanobacteria are sold as food, notably Arthrospira platensis (Spirulina) and others (Aphanizomenon flos-aquae).[249]
Some microalgae contain substances of high biological value, such as polyunsaturated fatty acids, amino acids, proteins, pigments, antioxidants, vitamins, and minerals.[250] Edible blue-green algae reduce the production of pro-inflammatory cytokines by inhibiting NF-κB pathway in macrophages and splenocytes.[251] Sulfate polysaccharides exhibit immunomodulatory, antitumor, antithrombotic, anticoagulant, anti-mutagenic, anti-inflammatory, antimicrobial, and even antiviral activity against HIV, herpes, and hepatitis.[252]
Health risks
Some cyanobacteria can produce neurotoxins, cytotoxins, endotoxins, and hepatotoxins (e.g., the microcystin-producing bacteria genus microcystis), which are collectively known as cyanotoxins.
Specific toxins include anatoxin-a, guanitoxin, aplysiatoxin, cyanopeptolin, cylindrospermopsin, domoic acid, nodularin R (from Nodularia), neosaxitoxin, and saxitoxin. Cyanobacteria reproduce explosively under certain conditions. This results in algal blooms which can become harmful to other species and pose a danger to humans and animals if the cyanobacteria involved produce toxins. Several cases of human poisoning have been documented, but a lack of knowledge prevents an accurate assessment of the risks,[253][254][255][256] and research by Linda Lawton, FRSE at Robert Gordon University, Aberdeen and collaborators has 30 years of examining the phenomenon and methods of improving water safety.[257]
Recent studies suggest that significant exposure to high levels of cyanobacteria producing toxins such as BMAA can cause amyotrophic lateral sclerosis (ALS). People living within half a mile of cyanobacterially contaminated lakes have had a 2.3 times greater risk of developing ALS than the rest of the population; people around New Hampshire's Lake Mascoma had an up to 25 times greater risk of ALS than the expected incidence.[258] BMAA from desert crusts found throughout Qatar might have contributed to higher rates of ALS in Gulf War veterans.[254][259]
Chemical control
Several chemicals can eliminate cyanobacterial blooms from smaller water-based systems such as swimming pools. They include calcium hypochlorite, copper sulphate, cupricide, and simazine.[260] The calcium hypochlorite amount needed varies depending on the cyanobacteria bloom, and treatment is needed periodically. According to the Department of Agriculture Australia, a rate of 12 g of 70% material in 1000 L of water is often effective to treat a bloom.[260] Copper sulfate is also used commonly, but no longer recommended by the Australian Department of Agriculture, as it kills livestock, crustaceans, and fish.[260] Cupricide is a chelated copper product that eliminates blooms with lower toxicity risks than copper sulfate. Dosage recommendations vary from 190 mL to 4.8 L per 1000 m2.[260] Ferric alum treatments at the rate of 50 mg/L will reduce algae blooms.[260][261] Simazine, which is also a herbicide, will continue to kill blooms for several days after an application. Simazine is marketed at different strengths (25, 50, and 90%), the recommended amount needed for one cubic meter of water per product is 25% product 8 mL; 50% product 4 mL; or 90% product 2.2 mL.[260]
Climate change
Climate change is likely to increase the frequency, intensity and duration of cyanobacterial blooms in many eutrophic lakes, reservoirs and estuaries.[262][31] Bloom-forming cyanobacteria produce a variety of neurotoxins, hepatotoxins and dermatoxins, which can be fatal to birds and mammals (including waterfowl, cattle and dogs) and threaten the use of waters for recreation, drinking water production, agricultural irrigation and fisheries.[31] Toxic cyanobacteria have caused major water quality problems, for example in Lake Taihu (China), Lake Erie (USA), Lake Okeechobee (USA), Lake Victoria (Africa) and the Baltic Sea.[31][263][264][265]
Climate change favours cyanobacterial blooms both directly and indirectly.[31] Many bloom-forming cyanobacteria can grow at relatively high temperatures.[266] Increased thermal stratification of lakes and reservoirs enables buoyant cyanobacteria to float upwards and form dense surface blooms, which gives them better access to light and hence a selective advantage over nonbuoyant phytoplankton organisms.[267][268] Protracted droughts during summer increase water residence times in reservoirs, rivers and estuaries, and these stagnant warm waters can provide ideal conditions for cyanobacterial bloom development.[269][265]
The capacity of the harmful cyanobacterial genus Microcystis to adapt to elevated CO2 levels was demonstrated in both laboratory and field experiments.[270] Microcystis spp. take up CO2 and HCO3− and accumulate inorganic carbon in carboxysomes, and strain competitiveness was found to depend on the concentration of inorganic carbon. As a result, climate change and increased CO2 levels are expected to affect the strain composition of cyanobacterial blooms.[270][265]
Gallery
 Cyanobacteria activity turns Coatepeque Caldera lake a turquoise color
Cyanobacteria activity turns Coatepeque Caldera lake a turquoise color Cyanobacterial bloom near Fiji
Cyanobacterial bloom near Fiji Cyanobacteria in Lake Köyliö.
Cyanobacteria in Lake Köyliö.- Video – Oscillatoria and Gleocapsa – with oscillatory movement as filaments of Oscillatoria orient towards light
See also
- Archean Eon
- Bacterial phyla, other major lineages of Bacteria
- Biodiesel
- Cyanobiont
- Endosymbiotic theory
- Geological history of oxygen
- Hypolith
Notes
- Botanists restrict the name algae to eukaryotes, which does not extend to cyanobacteria, which are prokaryotes. However, the common name blue-green algae continues to be used synonymously with cyanobacteria outside of the biological sciences.
References
- Komárek J, Kaštovský J, Mareš J, Johansen JR (2014). "Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach" (PDF). Preslia. 86: 295–335.
- Silva PC, Moe RL (December 2019). "Cyanophyceae". AccessScience. McGraw-Hill Education. doi:10.1036/1097-8542.175300. Retrieved 21 April 2011.
- Oren A (September 2004). "A proposal for further integration of the cyanobacteria under the Bacteriological Code". International Journal of Systematic and Evolutionary Microbiology. 54 (Pt 5): 1895–902. doi:10.1099/ijs.0.03008-0. PMID 15388760.
- Sinha, Rajeshwar P.; Häder, Donat-P. (2008). "UV-protectants in cyanobacteria" (PDF). Plant Science. 174 (3): 278–89. doi:10.1016/j.plantsci.2007.12.004. Archived from the original (PDF) on 15 April 2021.
- "cyan". Online Etymology Dictionary. Retrieved 21 January 2018.
- "Henry George Liddell, Robert Scott, A Greek-English Lexicon, κύα^νος". www.perseus.tufts.edu. Retrieved 21 January 2018.
- "Life History and Ecology of Cyanobacteria". University of California Museum of Paleontology. Archived from the original on 19 September 2012. Retrieved 17 July 2012.
- "Taxonomy Browser – Cyanobacteria". National Center for Biotechnology Information. NCBI:txid1117. Retrieved 12 April 2018.
- Allaby, M, ed. (1992). "Algae". The Concise Dictionary of Botany. Oxford: Oxford University Press.
- Stal LJ, Cretoiu MS (2016). The Marine Microbiome: An Untapped Source of Biodiversity and Biotechnological Potential. Springer. ISBN 978-3319330006.
- Monchamp, Marie-Eve; Spaak, Piet; Pomati, Francesco (27 July 2019). "Long Term Diversity and Distribution of Non-photosynthetic Cyanobacteria in Peri-Alpine Lakes". Frontiers in Microbiology. 9: 3344. doi:10.3389/fmicb.2018.03344. PMC 6340189. PMID 30692982.
- Liberton M, Pakrasi HB (2008). "Chapter 10. Membrane Systems in Cyanobacteria". In Herrero A, Flore E (eds.). The Cyanobacteria: Molecular Biology, Genomics, and Evolution. Norwich, United Kingdom: Horizon Scientific Press. pp. 217–87. ISBN 978-1-904455-15-8.
- Liberton M, Page LE, O'Dell WB, O'Neill H, Mamontov E, Urban VS, Pakrasi HB (February 2013). "Organization and flexibility of cyanobacterial thylakoid membranes examined by neutron scattering". The Journal of Biological Chemistry. 288 (5): 3632–40. doi:10.1074/jbc.M112.416933. PMC 3561581. PMID 23255600.
- Whitton BA, ed. (2012). "The fossil record of cyanobacteria". Ecology of Cyanobacteria II: Their Diversity in Space and Time. Springer Science & Business Media. pp. 17–. ISBN 978-94-007-3855-3.
- Basic Biology (18 March 2016). "Bacteria".
- Pathak, Jainendra; Rajneesh; Maurya, Pankaj K.; Singh, Shailendra P.; Häder, Donat-P.; Sinha, Rajeshwar P. (2018). "Cyanobacterial Farming for Environment Friendly Sustainable Agriculture Practices: Innovations and Perspectives". Frontiers in Environmental Science. 6. doi:10.3389/fenvs.2018.00007. ISSN 2296-665X.
- Morrison, Jim (11 January 2016). "Living Bacteria Are Riding Earth's Air Currents". Smithsonian Magazine. Retrieved 10 August 2022.
- Whitton, Brian A.; Potts, Malcolm (2012). "Introduction to the Cyanobacteria". Ecology of Cyanobacteria II. pp. 1–13. doi:10.1007/978-94-007-3855-3_1. ISBN 978-94-007-3854-6.
- Tamulonis, Carlos; Postma, Marten; Kaandorp, Jaap (2011). "Modeling Filamentous Cyanobacteria Reveals the Advantages of Long and Fast Trichomes for Optimizing Light Exposure". PLOS ONE. 6 (7): e22084. Bibcode:2011PLoSO...622084T. doi:10.1371/journal.pone.0022084. PMC 3138769. PMID 21789215.
- Whitton, Brian A. (5 July 2012). Ecology of Cyanobacteria II: Their Diversity in Space and Time. Springer Science & Business Media. ISBN 9789400738553. Retrieved 15 February 2022 – via Google Books.
- Tamulonis, Carlos; Postma, Marten; Kaandorp, Jaap (2011). "Modeling Filamentous Cyanobacteria Reveals the Advantages of Long and Fast Trichomes for Optimizing Light Exposure". PLOS ONE. 6 (7): e22084. Bibcode:2011PLoSO...622084T. doi:10.1371/journal.pone.0022084. PMC 3138769. PMID 21789215.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Kenneth R. Weiss (30 July 2006). "A Primeval Tide of Toxins". Los Angeles Times. Archived from the original on 14 August 2006.
- Dodds WK, Gudder DA, Mollenhauer D (1995). "The ecology of 'Nostoc'". Journal of Phycology. 31: 2–18. doi:10.1111/j.0022-3646.1995.00002.x. S2CID 85011483.
- Aguilera, Anabella; Klemenčič, Marina; Sueldo, Daniela J.; Rzymski, Piotr; Giannuzzi, Leda; Martin, María Victoria (2021). "Cell Death in Cyanobacteria: Current Understanding and Recommendations for a Consensus on Its Nomenclature". Frontiers in Microbiology. 12: 631654. doi:10.3389/fmicb.2021.631654. PMC 7965980. PMID 33746925.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Whitton, Brian A. (5 July 2012). Ecology of Cyanobacteria II: Their Diversity in Space and Time. ISBN 9789400738553.
- Schopf, J.; Packer, B. (1987). "Early Archean (3.3-billion to 3.5-billion-year-old) microfossils from Warrawoona Group, Australia". Science. 237 (4810): 70–73. Bibcode:1987Sci...237...70S. doi:10.1126/science.11539686. PMID 11539686.
- Bullerjahn, George S.; Post, Anton F. (2014). "Physiology and molecular biology of aquatic cyanobacteria". Frontiers in Microbiology. 5: 359. doi:10.3389/fmicb.2014.00359. PMC 4099938. PMID 25076944.
- Tang, Weiyi; Wang, Seaver; Fonseca-Batista, Debany; Dehairs, Frank; Gifford, Scott; Gonzalez, Aridane G.; Gallinari, Morgane; Planquette, Hélène; Sarthou, Géraldine; Cassar, Nicolas (2019). "Revisiting the distribution of oceanic N2 fixation and estimating diazotrophic contribution to marine production". Nature Communications. 10 (1): 831. doi:10.1038/s41467-019-08640-0. PMC 6381160. PMID 30783106.
- Bláha, Luděk; Babica, Pavel; Maršálek, Blahoslav (2009). "Toxins produced in cyanobacterial water blooms - toxicity and risks". Interdisciplinary Toxicology. 2 (2): 36–41. doi:10.2478/v10102-009-0006-2. PMC 2984099. PMID 21217843.
- Paerl, Hans W.; Otten, Timothy G. (2013). "Harmful Cyanobacterial Blooms: Causes, Consequences, and Controls". Microbial Ecology. 65 (4): 995–1010. doi:10.1007/s00248-012-0159-y. PMID 23314096. S2CID 5718333.
- Huisman, Jef; Codd, Geoffrey A.; Paerl, Hans W.; Ibelings, Bas W.; Verspagen, Jolanda M. H.; Visser, Petra M. (2018). "Cyanobacterial blooms". Nature Reviews Microbiology. 16 (8): 471–483. doi:10.1038/s41579-018-0040-1. PMID 29946124. S2CID 49427202.
- Field, C. B.; Behrenfeld, M. J.; Randerson, J. T.; Falkowski, P. (1998). "Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components". Science. 281 (5374): 237–240. Bibcode:1998Sci...281..237F. doi:10.1126/science.281.5374.237. PMID 9657713.
- Zehr, Jonathan P. (2011). "Nitrogen fixation by marine cyanobacteria". Trends in Microbiology. 19 (4): 162–173. doi:10.1016/j.tim.2010.12.004. PMID 21227699.
- Thompson, A. W.; Foster, R. A.; Krupke, A.; Carter, B. J.; Musat, N.; Vaulot, D.; Kuypers, M. M. M.; Zehr, J. P. (2012). "Unicellular Cyanobacterium Symbiotic with a Single-Celled Eukaryotic Alga". Science. 337 (6101): 1546–1550. Bibcode:2012Sci...337.1546T. doi:10.1126/science.1222700. PMID 22997339. S2CID 7071725.
- Johnson, Z. I.; Zinser, E. R.; Coe, A.; McNulty, N. P.; Woodward, E. M.; Chisholm, S. W. (2006). "Niche Partitioning Among Prochlorococcus Ecotypes Along Ocean-Scale Environmental Gradients". Science. 311 (5768): 1737–1740. Bibcode:2006Sci...311.1737J. doi:10.1126/science.1118052. PMID 16556835. S2CID 3549275.
- Scanlan, D. J.; Ostrowski, M.; Mazard, S.; Dufresne, A.; Garczarek, L.; Hess, W. R.; Post, A. F.; Hagemann, M.; Paulsen, I.; Partensky, F. (2009). "Ecological Genomics of Marine Picocyanobacteria". Microbiology and Molecular Biology Reviews. 73 (2): 249–299. doi:10.1128/MMBR.00035-08. PMC 2698417. PMID 19487728.
- Flombaum, P.; Gallegos, J. L.; Gordillo, R. A.; Rincon, J.; Zabala, L. L.; Jiao, N.; Karl, D. M.; Li, W. K. W.; Lomas, M. W.; Veneziano, D.; Vera, C. S.; Vrugt, J. A.; Martiny, A. C. (2013). "Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus". Proceedings of the National Academy of Sciences. 110 (24): 9824–9829. Bibcode:2013PNAS..110.9824F. doi:10.1073/pnas.1307701110. PMC 3683724. PMID 23703908.
- Foster, Rachel A.; Kuypers, Marcel M M.; Vagner, Tomas; Paerl, Ryan W.; Musat, Niculina; Zehr, Jonathan P. (2011). "Nitrogen fixation and transfer in open ocean diatom–cyanobacterial symbioses". The ISME Journal. 5 (9): 1484–1493. doi:10.1038/ismej.2011.26. PMC 3160684. PMID 21451586.
- Villareal, Tracy A. (1990). "Laboratory Culture and Preliminary Characterization of the Nitrogen-Fixing Rhizosolenia-Richelia Symbiosis". Marine Ecology. 11 (2): 117–132. Bibcode:1990MarEc..11..117V. doi:10.1111/j.1439-0485.1990.tb00233.x.
- Janson; Wouters; Bergman; Carpenter (1999). "Host specificity in the Richelia-diatom symbiosis revealed by hetR gene sequence analysis". Environmental Microbiology. 1 (5): 431–438. doi:10.1046/j.1462-2920.1999.00053.x. PMID 11207763.
- Sánchez-Baracaldo, Patricia (2015). "Origin of marine planktonic cyanobacteria". Scientific Reports. 5: 17418. Bibcode:2015NatSR...517418S. doi:10.1038/srep17418. PMC 4665016. PMID 26621203.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Kettler GC, Martiny AC, Huang K, Zucker J, Coleman ML, Rodrigue S, Chen F, Lapidus A, Ferriera S, Johnson J, Steglich C, Church GM, Richardson P, Chisholm SW (December 2007). "Patterns and implications of gene gain and loss in the evolution of Prochlorococcus". PLOS Genetics. 3 (12): e231. doi:10.1371/journal.pgen.0030231. PMC 2151091. PMID 18159947.
- Nemiroff, R.; Bonnell, J., eds. (27 September 2006). "Earth from Saturn". Astronomy Picture of the Day. NASA.
- Partensky F, Hess WR, Vaulot D (March 1999). "Prochlorococcus, a marine photosynthetic prokaryote of global significance". Microbiology and Molecular Biology Reviews. 63 (1): 106–27. doi:10.1128/MMBR.63.1.106-127.1999. PMC 98958. PMID 10066832.
- "The Most Important Microbe You've Never Heard Of". npr.org.
- Claessen, Dennis; Rozen, Daniel E.; Kuipers, Oscar P.; Søgaard-Andersen, Lotte; Van Wezel, Gilles P. (2014). "Bacterial solutions to multicellularity: A tale of biofilms, filaments and fruiting bodies". Nature Reviews Microbiology. 12 (2): 115–124. doi:10.1038/nrmicro3178. hdl:11370/0db66a9c-72ef-4e11-a75d-9d1e5827573d. PMID 24384602. S2CID 20154495.
- Nürnberg, Dennis J.; Mariscal, Vicente; Parker, Jamie; Mastroianni, Giulia; Flores, Enrique; Mullineaux, Conrad W. (2014). "Branching and intercellular communication in the Section V cyanobacterium Mastigocladus laminosus, a complex multicellular prokaryote". Molecular Microbiology. 91 (5): 935–949. doi:10.1111/mmi.12506. hdl:10261/99110. PMID 24383541. S2CID 25479970.
- Herrero, Antonia; Stavans, Joel; Flores, Enrique (2016). "The multicellular nature of filamentous heterocyst-forming cyanobacteria". FEMS Microbiology Reviews. 40 (6): 831–854. doi:10.1093/femsre/fuw029. hdl:10261/140753. PMID 28204529.
- Risser DD, Chew WG, Meeks JC (April 2014). "Genetic characterization of the hmp locus, a chemotaxis-like gene cluster that regulates hormogonium development and motility in Nostoc punctiforme". Molecular Microbiology. 92 (2): 222–33. doi:10.1111/mmi.12552. PMID 24533832. S2CID 37479716.
- Khayatan B, Bains DK, Cheng MH, Cho YW, Huynh J, Kim R, Omoruyi OH, Pantoja AP, Park JS, Peng JK, Splitt SD, Tian MY, Risser DD (May 2017). "A Putative O-Linked β-N-Acetylglucosamine Transferase Is Essential for Hormogonium Development and Motility in the Filamentous Cyanobacterium Nostoc punctiforme". Journal of Bacteriology. 199 (9): e00075–17. doi:10.1128/JB.00075-17. PMC 5388816. PMID 28242721.
- Esteves-Ferreira, Alberto A.; Cavalcanti, João Henrique Frota; Vaz, Marcelo Gomes Marçal Vieira; Alvarenga, Luna V.; Nunes-Nesi, Adriano; Araújo, Wagner L. (2017). "Cyanobacterial nitrogenases: Phylogenetic diversity, regulation and functional predictions". Genetics and Molecular Biology. 40 (1 suppl 1): 261–275. doi:10.1590/1678-4685-GMB-2016-0050. PMC 5452144. PMID 28323299.
- Meeks JC, Elhai J, Thiel T, Potts M, Larimer F, Lamerdin J, Predki P, Atlas R (2001). "An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium". Photosynthesis Research. 70 (1): 85–106. doi:10.1023/A:1013840025518. PMID 16228364. S2CID 8752382.
- Golden JW, Yoon HS (December 1998). "Heterocyst formation in Anabaena". Current Opinion in Microbiology. 1 (6): 623–9. doi:10.1016/s1369-5274(98)80106-9. PMID 10066546.
- Fay P (June 1992). "Oxygen relations of nitrogen fixation in cyanobacteria". Microbiological Reviews. 56 (2): 340–73. doi:10.1128/MMBR.56.2.340-373.1992. PMC 372871. PMID 1620069.
- Singh. Text Book of Botany Diversity of Microbes And Cryptogams. Rastogi Publications. ISBN 978-8171338894.
- "Differences between Bacteria and Cyanobacteria". Microbiology Notes. 29 October 2015. Retrieved 21 January 2018.
- Walsby AE (March 1994). "Gas vesicles". Microbiological Reviews. 58 (1): 94–144. doi:10.1128/MMBR.58.1.94-144.1994. PMC 372955. PMID 8177173.
-
Sims GK, Dunigan EP (1984). "Diurnal and seasonal variations in nitrogenase activity C
2H
2 reduction) of rice roots". Soil Biology and Biochemistry. 16: 15–18. doi:10.1016/0038-0717(84)90118-4. - Bocchi S, Malgioglio A (2010). "Azolla-Anabaena as a Biofertilizer for Rice Paddy Fields in the Po Valley, a Temperate Rice Area in Northern Italy". International Journal of Agronomy. 2010: 1–5. doi:10.1155/2010/152158.
- Huokko, Tuomas; Ni, Tao; Dykes, Gregory F.; Simpson, Deborah M.; Brownridge, Philip; Conradi, Fabian D.; Beynon, Robert J.; Nixon, Peter J.; Mullineaux, Conrad W.; Zhang, Peijun; Liu, Lu-Ning (2021). "Probing the biogenesis pathway and dynamics of thylakoid membranes". Nature Communications. 12 (1): 3475. Bibcode:2021NatCo..12.3475H. doi:10.1038/s41467-021-23680-1. PMC 8190092. PMID 34108457.
- Kerfeld CA, Heinhorst S, Cannon GC (2010). "Bacterial microcompartments". Annual Review of Microbiology. 64 (1): 391–408. doi:10.1146/annurev.micro.112408.134211. PMC 6022854. PMID 20825353.
- Rae, Benjamin D.; Long, Benedict M.; Badger, Murray R.; Price, G. Dean (September 2013). "Functions, Compositions, and Evolution of the Two Types of Carboxysomes: Polyhedral Microcompartments That Facilitate CO 2 Fixation in Cyanobacteria and Some Proteobacteria". Microbiology and Molecular Biology Reviews. 77 (3): 357–379. doi:10.1128/MMBR.00061-12. ISSN 1092-2172. PMC 3811607. PMID 24006469.
- Long BM, Badger MR, Whitney SM, Price GD (October 2007). "Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA". The Journal of Biological Chemistry. 282 (40): 29323–35. doi:10.1074/jbc.M703896200. PMID 17675289.
- Vothknecht UC, Westhoff P (December 2001). "Biogenesis and origin of thylakoid membranes". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 1541 (1–2): 91–101. doi:10.1016/S0167-4889(01)00153-7. PMID 11750665.
- Sobiechowska-Sasim M, Stoń-Egiert J, Kosakowska A (February 2014). "Quantitative analysis of extracted phycobilin pigments in cyanobacteria – an assessment of spectrophotometric and spectrofluorometric methods". J Appl Phycol. 26 (5): 2065–74. doi:10.1007/s10811-014-0244-3. PMC 4200375. PMID 25346572.
- Pisciotta JM, Zou Y, Baskakov IV (May 2010). Yang C (ed.). "Light-dependent electrogenic activity of cyanobacteria". PLOS ONE. 5 (5): e10821. Bibcode:2010PLoSO...510821P. doi:10.1371/journal.pone.0010821. PMC 2876029. PMID 20520829.
- Vermaas WF (2001). "Photosynthesis and Respiration in Cyanobacteria". Photosynthesis and Respiration in Cyanobacteria. eLS. John Wiley & Sons, Ltd. doi:10.1038/npg.els.0001670. ISBN 978-0-470-01590-2. S2CID 19016706.
- Armstronf JE (2015). How the Earth Turned Green: A Brief 3.8-Billion-Year History of Plants. The University of Chicago Press. ISBN 978-0-226-06977-7.
- Klatt JM, de Beer D, Häusler S, Polerecky L (2016). "Cyanobacteria in Sulfidic Spring Microbial Mats Can Perform Oxygenic and Anoxygenic Photosynthesis Simultaneously during an Entire Diurnal Period". Frontiers in Microbiology. 7: 1973. doi:10.3389/fmicb.2016.01973. PMC 5156726. PMID 28018309.
- Grossman AR, Schaefer MR, Chiang GG, Collier JL (September 1993). "The phycobilisome, a light-harvesting complex responsive to environmental conditions". Microbiological Reviews. 57 (3): 725–49. doi:10.1128/MMBR.57.3.725-749.1993. PMC 372933. PMID 8246846.
- "Colors from bacteria | Causes of Color". www.webexhibits.org. Retrieved 22 January 2018.
- Garcia-Pichel F (2009). "Cyanobacteria". In Schaechter M (ed.). Encyclopedia of Microbiology (third ed.). pp. 107–24. doi:10.1016/B978-012373944-5.00250-9. ISBN 978-0-12-373944-5.
- Kehoe DM (May 2010). "Chromatic adaptation and the evolution of light color sensing in cyanobacteria". Proceedings of the National Academy of Sciences of the United States of America. 107 (20): 9029–30. Bibcode:2010PNAS..107.9029K. doi:10.1073/pnas.1004510107. PMC 2889117. PMID 20457899.
- Kehoe DM, Gutu A (2006). "Responding to color: the regulation of complementary chromatic adaptation". Annual Review of Plant Biology. 57: 127–50. doi:10.1146/annurev.arplant.57.032905.105215. PMID 16669758.
- Palenik B, Haselkorn R (January 1992). "Multiple evolutionary origins of prochlorophytes, the chlorophyll b-containing prokaryotes". Nature. 355 (6357): 265–67. Bibcode:1992Natur.355..265P. doi:10.1038/355265a0. PMID 1731224. S2CID 4244829.
- Urbach E, Robertson DL, Chisholm SW (January 1992). "Multiple evolutionary origins of prochlorophytes within the cyanobacterial radiation". Nature. 355 (6357): 267–70. Bibcode:1992Natur.355..267U. doi:10.1038/355267a0. PMID 1731225. S2CID 2011379.
- Cohen Y, Jørgensen BB, Revsbech NP, Poplawski R (February 1986). "Adaptation to Hydrogen Sulfide of Oxygenic and Anoxygenic Photosynthesis among Cyanobacteria". Applied and Environmental Microbiology. 51 (2): 398–407. Bibcode:1986ApEnM..51..398C. doi:10.1128/AEM.51.2.398-407.1986. PMC 238881. PMID 16346996.
- Blankenship RE (2014). Molecular Mechanisms of Photosynthesis. Wiley-Blackwell. pp. 147–73. ISBN 978-1-4051-8975-0.
- Och LM, Shields-Zhou GA (January 2012). "The Neoproterozoic oxygenation event: Environmental perturbations and biogeochemical cycling". Earth-Science Reviews. 110 (1–4): 26–57. Bibcode:2012ESRv..110...26O. doi:10.1016/j.earscirev.2011.09.004.
- Adams DG, Bergman B, Nierzwicki-Bauer SA, Duggan PS, Rai AN, Schüßler A (2013). The Prokaryotes. Springer, Berlin, Heidelberg. pp. 359–400. doi:10.1007/978-3-642-30194-0_17. ISBN 978-3-642-30193-3.
- Smith A (1973). "Synthesis of metabolic intermediates". In Carr NG, Whitton BA (eds.). The Biology of Blue-green Algae. University of California Press. pp. 30–. ISBN 978-0-520-02344-4.
- Jangoux M (1987). "Diseases of Echinodermata. I. Agents microorganisms and protistans". Dis. Aquat. Org. 2: 147–62. doi:10.3354/dao002147.
- Kinne O, ed. (1980). Diseases of Marine Animals (PDF). Vol. 1. Chichester, UK: John Wiley & Sons. ISBN 978-0-471-99584-5.
- Kristiansen A (1964). "Sarcinastrum urosporae, a Colourless Parasitic Blue-green Alga" (PDF). Phycologia. 4 (1): 19–22. doi:10.2216/i0031-8884-4-1-19.1. Archived from the original (PDF) on 6 January 2015.
- Mazard S, Penesyan A, Ostrowski M, Paulsen IT, Egan S (2016). "Tiny microbes with a big impact: the role of cyanobacteria and their metabolites in shaping our future". Marine Drugs. 14 (5): 97. doi:10.3390/md14050097. PMC 4882571. PMID 27196915.
- de los Ríos A, Grube M, Sancho LG, Ascaso C (February 2007). "Ultrastructural and genetic characteristics of endolithic cyanobacterial biofilms colonizing Antarctic granite rocks". FEMS Microbiology Ecology. 59 (2): 386–95. doi:10.1111/j.1574-6941.2006.00256.x. PMID 17328119.
- Vaughan T (2011). Mammalogy. Jones and Barlett. p. 21. ISBN 978-0763762995.
- Schultz, Nora (30 August 2009). "Photosynthetic viruses keep world's oxygen levels up". New Scientist.
- Jöhnk KD, Huisman J, Sharples J, Sommeijer B, Visser PM, Stroom JM (1 March 2008). "Summer heatwaves promote blooms of harmful cyanobacteria". Global Change Biology. 14 (3): 495–512. Bibcode:2008GCBio..14..495J. doi:10.1111/j.1365-2486.2007.01510.x. S2CID 54079634.
- "Linda Lawton – 11th International Conference on Toxic Cyanobacteria". Retrieved 25 June 2021.
- Paerl HW, Paul VJ (April 2012). "Climate change: links to global expansion of harmful cyanobacteria". Water Research. 46 (5): 1349–63. doi:10.1016/j.watres.2011.08.002. PMID 21893330.
- Thomas AD, Dougill AJ (15 March 2007). "Spatial and temporal distribution of cyanobacterial soil crusts in the Kalahari: Implications for soil surface properties". Geomorphology. 85 (1): 17–29. Bibcode:2007Geomo..85...17T. doi:10.1016/j.geomorph.2006.03.029.
- Belnap J, Gardner JS (1993). "Soil Microstructure in Soils of the Colorado Plateau: The Role of the Cyanobacterium Microcoleus Vaginatus". The Great Basin Naturalist. 53 (1): 40–47. JSTOR 41712756.
- Nadis S (December 2003). "The cells that rule the seas" (PDF). Scientific American. 289 (6): 52–53. Bibcode:2003SciAm.289f..52N. doi:10.1038/scientificamerican1203-52. PMID 14631732. Archived from the original (PDF) on 19 April 2014. Retrieved 19 April 2014.
- Walsh PJ, Smith S, Fleming L, Solo-Gabriele H, Gerwick WH, eds. (2 September 2011). "Cyanobacteria and cyanobacterial toxins". Oceans and Human Health: Risks and Remedies from the Seas. Academic Press. pp. 271–96. ISBN 978-0-08-087782-2.
- Lee, Sang-Moo; Ryu, Choong-Min (4 February 2021). "Algae as New Kids in the Beneficial Plant Microbiome". Frontiers in Plant Science. Frontiers Media SA. 12: 599742. doi:10.3389/fpls.2021.599742. ISSN 1664-462X. PMC 7889962. PMID 33613596.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Kim, Miran; Choi, Dong Han; Park, Myung Gil (2021). "Cyanobiont genetic diversity and host specificity of cyanobiont-bearing dinoflagellate Ornithocercus in temperate coastal waters". Scientific Reports. 11 (1): 9458. Bibcode:2021NatSR..11.9458K. doi:10.1038/s41598-021-89072-z. PMC 8097063. PMID 33947914.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Gantar, M.; Elhai, J. (1999). "Colonization of wheatpara-nodules by the N2-fixing cyanobacterium Nostocsp. Strain 2S9B". New Phytologist. 141 (3): 373–379. doi:10.1046/j.1469-8137.1999.00352.x.
- Gantar, M. (2000). "Mechanical damage of roots provides enhanced colonization of the wheat endorhizosphere by the dinitrogen-fixing cyanobacterium Nostoc sp. Strain 2S9B". Biology and Fertility of Soils. 32 (3): 250–255. doi:10.1007/s003740000243. S2CID 7590731.
- Treves, Haim; Raanan, Hagai; Kedem, Isaac; Murik, Omer; Keren, Nir; Zer, Hagit; Berkowicz, Simon M.; Giordano, Mario; Norici, Alessandra; Shotland, Yoram; Ohad, Itzhak; Kaplan, Aaron (2016). "The mechanisms whereby the green alga Chlorella ohadii , isolated from desert soil crust, exhibits unparalleled photodamage resistance". New Phytologist. 210 (4): 1229–1243. doi:10.1111/nph.13870. PMID 26853530.
- Zhu, Huan; Li, Shuyin; Hu, Zhengyu; Liu, Guoxiang (2018). "Molecular characterization of eukaryotic algal communities in the tropical phyllosphere based on real-time sequencing of the 18S rDNA gene". BMC Plant Biology. 18 (1): 365. doi:10.1186/s12870-018-1588-7. PMC 6299628. PMID 30563464.
- Krings, Michael; Hass, Hagen; Kerp, Hans; Taylor, Thomas N.; Agerer, Reinhard; Dotzler, Nora (2009). "Endophytic cyanobacteria in a 400-million-yr-old land plant: A scenario for the origin of a symbiosis?". Review of Palaeobotany and Palynology. 153 (1–2): 62–69. doi:10.1016/j.revpalbo.2008.06.006.
- Karthikeyan, N.; Prasanna, R.; Sood, A.; Jaiswal, P.; Nayak, S.; Kaushik, B. D. (2009). "Physiological characterization and electron microscopic investigation of cyanobacteria associated with wheat rhizosphere". Folia Microbiologica. 54 (1): 43–51. doi:10.1007/s12223-009-0007-8. PMID 19330544. S2CID 23420342.
- Babu, Santosh; Prasanna, Radha; Bidyarani, Ngangom; Singh, Rajendra (2015). "Analysing the colonisation of inoculated cyanobacteria in wheat plants using biochemical and molecular tools". Journal of Applied Phycology. 27: 327–338. doi:10.1007/s10811-014-0322-6. S2CID 17353123.
- Bidyarani, Ngangom; Prasanna, Radha; Chawla, Gautam; Babu, Santosh; Singh, Rajendra (2015). "Deciphering the factors associated with the colonization of rice plants by cyanobacteria". Journal of Basic Microbiology. 55 (4): 407–419. doi:10.1002/jobm.201400591. PMID 25515189. S2CID 5401526.
- Gantar, M.; Kerby, N. W.; Rowell, P. (1991). "Colonization of wheat (Triticum vulgare L.) by N2-fixing cyanobacteria: II. An ultrastructural study". New Phytologist. 118 (3): 485–492. doi:10.1111/j.1469-8137.1991.tb00031.x.
- Ahmed, Mehboob; Stal, Lucas J.; Hasnain, Shahida (2010). "Association of non-heterocystous cyanobacteria with crop plants". Plant and Soil. 336 (1–2): 363–375. doi:10.1007/s11104-010-0488-x. S2CID 21309970.
- Hussain, Anwar; Hamayun, Muhammad; Shah, Syed Tariq (2013). "Root Colonization and Phytostimulation by Phytohormones Producing Entophytic Nostoc sp. AH-12". Current Microbiology. 67 (5): 624–630. doi:10.1007/s00284-013-0408-4. PMID 23794014. S2CID 14704537.
- Hussain, Anwar; Shah, Syed T.; Rahman, Hazir; Irshad, Muhammad; Iqbal, Amjad (2015). "Effect of IAA on in vitro growth and colonization of Nostoc in plant roots". Frontiers in Plant Science. 6: 46. doi:10.3389/fpls.2015.00046. PMC 4318279. PMID 25699072.
- Capone, D. G. (1997). "Trichodesmium, a Globally Significant Marine Cyanobacterium". Science. 276 (5316): 1221–1229. doi:10.1126/science.276.5316.1221.
- Falkowski, P. G. (1998). "Biogeochemical Controls and Feedbacks on Ocean Primary Production". Science. 281 (5374): 200–206. doi:10.1126/science.281.5374.200. PMID 9660741.
- Hutchins, D. A.; Fu, F.-X.; Zhang, Y.; Warner, M. E.; Feng, Y.; Portune, K.; Bernhardt, P. W.; Mulholland, M. R. (2007). "CO2 control of Trichodesmium N2 fixation, photosynthesis, growth rates, and elemental ratios: Implications for past, present, and future ocean biogeochemistry". Limnology and Oceanography. 52 (4): 1293–1304. Bibcode:2007LimOc..52.1293H. doi:10.4319/lo.2007.52.4.1293. S2CID 55606811.
- Huang, Sijun; Wilhelm, Steven W.; Harvey, H Rodger; Taylor, Karen; Jiao, Nianzhi; Chen, Feng (2012). "Novel lineages of Prochlorococcus and Synechococcus in the global oceans". The ISME Journal. 6 (2): 285–297. doi:10.1038/ismej.2011.106. PMC 3260499. PMID 21955990.
- Partensky, F.; Hess, W. R.; Vaulot, D. (1999). "Prochlorococcus , a Marine Photosynthetic Prokaryote of Global Significance". Microbiology and Molecular Biology Reviews. 63 (1): 106–127. doi:10.1128/MMBR.63.1.106-127.1999. PMC 98958. PMID 10066832.
- Srivastava, Ashish Kumar; Rai, Amar Nath; Neilan, Brett A. (March 2013). Stress Biology of Cyanobacteria: Molecular Mechanisms to Cellular Responses. ISBN 9781466504783.
- Zehr, J. P.; Bench, S. R.; Carter, B. J.; Hewson, I.; Niazi, F.; Shi, T.; Tripp, H. J.; Affourtit, J. P. (2008). "Globally Distributed Uncultivated Oceanic N2-Fixing Cyanobacteria Lack Oxygenic Photosystem II". Science. 322 (5904): 1110–1112. Bibcode:2008Sci...322.1110Z. doi:10.1126/science.1165340. PMID 19008448. S2CID 206516012.
- Decelle, Johan; Colin, Sébastien; Foster, Rachel A. (2015). "Photosymbiosis in Marine Planktonic Protists". Marine Protists. pp. 465–500. doi:10.1007/978-4-431-55130-0_19. ISBN 978-4-431-55129-4.
- Foster, Rachel A.; Zehr, Jonathan P. (2019). "Diversity, Genomics, and Distribution of Phytoplankton-Cyanobacterium Single-Cell Symbiotic Associations". Annual Review of Microbiology. 73: 435–456. doi:10.1146/annurev-micro-090817-062650. PMID 31500535. S2CID 202414294.
- Mullineaux, Conrad W.; Wilde, Annegret (2021). "The social life of cyanobacteria". eLife. 10. doi:10.7554/eLife.70327. PMC 8208810. PMID 34132636.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Kamennaya, Nina A.; Zemla, Marcin; Mahoney, Laura; Chen, Liang; Holman, Elizabeth; Holman, Hoi-Ying; Auer, Manfred; Ajo-Franklin, Caroline M.; Jansson, Christer (2018). "High pCO2-induced exopolysaccharide-rich ballasted aggregates of planktonic cyanobacteria could explain Paleoproterozoic carbon burial". Nature Communications. 9 (1): 2116. Bibcode:2018NatCo...9.2116K. doi:10.1038/s41467-018-04588-9. PMC 5974010. PMID 29844378.
- Maeda, Kaisei; Okuda, Yukiko; Enomoto, Gen; Watanabe, Satoru; Ikeuchi, Masahiko (2021). "Biosynthesis of a sulfated exopolysaccharide, synechan, and bloom formation in the model cyanobacterium Synechocystis sp. Strain PCC 6803". eLife. 10. doi:10.7554/eLife.66538. PMC 8205485. PMID 34127188.
- Sim, Min Sub; Liang, Biqing; Petroff, Alexander P.; Evans, Alexander; Klepac-Ceraj, Vanja; Flannery, David T.; Walter, Malcolm R.; Bosak, Tanja (2012). "Oxygen-Dependent Morphogenesis of Modern Clumped Photosynthetic Mats and Implications for the Archean Stromatolite Record". Geosciences. 2 (4): 235–259. Bibcode:2012Geosc...2..235S. doi:10.3390/geosciences2040235.
 Material was copied from this source, which is available under a Creative Commons Attribution 3.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 3.0 International License. - Conradi, Fabian D.; Zhou, Rui-Qian; Oeser, Sabrina; Schuergers, Nils; Wilde, Annegret; Mullineaux, Conrad W. (2019). "Factors Controlling Floc Formation and Structure in the Cyanobacterium Synechocystis sp. Strain PCC 6803". Journal of Bacteriology. 201 (19). doi:10.1128/JB.00344-19. PMC 6755745. PMID 31262837.
- Enomoto, Gen; Ikeuchi, Masahiko (2020). "Blue-/Green-Light-Responsive Cyanobacteriochromes Are Cell Shade Sensors in Red-Light Replete Niches". iScience. 23 (3): 100936. Bibcode:2020iSci...23j0936E. doi:10.1016/j.isci.2020.100936. PMC 7063230. PMID 32146329.
- Galluzzi, L.; et al. (2015). "Essential versus accessory aspects of cell death: Recommendations of the NCCD 2015". Cell Death & Differentiation. 22 (1): 58–73. doi:10.1038/cdd.2014.137. PMC 4262782. PMID 25236395.
- Allen, Rey; Rittmann, Bruce E.; Curtiss, Roy (2019). "Axenic Biofilm Formation and Aggregation by Synechocystis sp. Strain PCC 6803 Are Induced by Changes in Nutrient Concentration and Require Cell Surface Structures". Applied and Environmental Microbiology. 85 (7). Bibcode:2019ApEnM..85E2192A. doi:10.1128/AEM.02192-18. PMC 6585507. PMID 30709828.
- Schuergers, Nils; Wilde, Annegret (2015). "Appendages of the Cyanobacterial Cell". Life. 5 (1): 700–715. doi:10.3390/life5010700. PMC 4390875. PMID 25749611.
- Khayatan, Behzad; Meeks, John C.; Risser, Douglas D. (2015). "Evidence that a modified type IV pilus-like system powers gliding motility and polysaccharide secretion in filamentous cyanobacteria". Molecular Microbiology. 98 (6): 1021–1036. doi:10.1111/mmi.13205. PMID 26331359. S2CID 8749419.
- Adams, David. W.; Stutzmann, Sandrine; Stoudmann, Candice; Blokesch, Melanie (2019). "DNA-uptake pili of Vibrio cholerae are required for chitin colonization and capable of kin recognition via sequence-specific self-interaction". Nature Microbiology. 4 (9): 1545–1557. doi:10.1038/s41564-019-0479-5. PMC 6708440. PMID 31182799.
- Kromkamp, Jacco; Walsby, Anthony E. (1990). "A computer model of buoyancy and vertical migration in cyanobacteria". Journal of Plankton Research. 12: 161–183. doi:10.1093/plankt/12.1.161.
- Aguilo-Ferretjans, Maria del Mar; Bosch, Rafael; Puxty, Richard J.; Latva, Mira; Zadjelovic, Vinko; Chhun, Audam; Sousoni, Despoina; Polin, Marco; Scanlan, David J.; Christie-Oleza, Joseph A. (2021). "Pili allow dominant marine cyanobacteria to avoid sinking and evade predation". Nature Communications. 12 (1): 1857. Bibcode:2021NatCo..12.1857A. doi:10.1038/s41467-021-22152-w. PMC 7994388. PMID 33767153.
- Agustí, S. (2004). "Viability and niche segregation of Prochlorococcus and Synechococcus cells across the Central Atlantic Ocean." Accessed: 30 July 2021).
- Agusti, Susana; Alou, EVA; Hoyer, Mark V.; Frazer, Thomas K.; Canfield, Daniel E. (2006). "Cell death in lake phytoplankton communities". Freshwater Biology. 51 (8): 1496–1506. doi:10.1111/j.1365-2427.2006.01584.x.
- Franklin, Daniel J.; Brussaard, Corina P.D.; Berges, John A. (2006). "What is the role and nature of programmed cell death in phytoplankton ecology?". European Journal of Phycology. 41: 1–14. doi:10.1080/09670260500505433. S2CID 53599616.
- Sigee, D. C.; Selwyn, A.; Gallois, P.; Dean, A. P. (2007). "Patterns of cell death in freshwater colonial cyanobacteria during the late summer bloom". Phycologia. 46 (3): 284–292. doi:10.2216/06-69.1. S2CID 86268392.
- Berman-Frank, Ilana; Bidle, Kay D.; Haramaty, Liti; Falkowski, Paul G. (2004). "The demise of the marine cyanobacterium, Trichodesmium SPP., via an autocatalyzed cell death pathway". Limnology and Oceanography. 49 (4): 997–1005. Bibcode:2004LimOc..49..997B. doi:10.4319/lo.2004.49.4.0997.
- Hu, Chenlin; Rzymski, Piotr (2019). "Programmed Cell Death-Like and Accompanying Release of Microcystin in Freshwater Bloom-Forming Cyanobacterium Microcystis: From Identification to Ecological Relevance". Toxins. 11 (12): 706. doi:10.3390/toxins11120706. PMC 6950475. PMID 31817272.
- Meeks, John C.; Elhai, Jeff; Thiel, Teresa; Potts, Malcolm; Larimer, Frank; Lamerdin, Jane; Predki, Paul; Atlas, Ronald (2001). "An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium". Photosynthesis Research. 70 (1): 85–106. doi:10.1023/A:1013840025518. PMID 16228364. S2CID 8752382.
- Suttle, Curtis A. (1 January 2000). "Cyanophages and Their Role in the Ecology of Cyanobacteria". In Whitton, Brian A.; Potts, Malcolm (eds.). The Ecology of Cyanobacteria. Springer Netherlands. pp. 563–589. doi:10.1007/0-306-46855-7_20. ISBN 9780792347354.
- Suttle, Curtis A.; Chan, Amy M. (1993). "Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, . morphology, cross-infectivity and growth characteristics". Marine Ecology Progress Series. 92: 99–109. Bibcode:1993MEPS...92...99S. doi:10.3354/meps092099.
- Fokine, Andrei; Rossmann, Michael G. (1 January 2014). "Molecular architecture of tailed double-stranded DNA phages". Bacteriophage. 4 (1): e28281. doi:10.4161/bact.28281. PMC 3940491. PMID 24616838.
- Proctor, Lita M.; Fuhrman, Jed A. (1990). "Viral mortality of marine bacteria and cyanobacteria". Nature. 343 (6253): 60–62. Bibcode:1990Natur.343...60P. doi:10.1038/343060a0. S2CID 4336344.
- Sarma TA. 'Cyanophages' in Handbook of Cyanobacteria (CRC Press; 2012) (ISBN 1466559411)
- Duggan, Paula S.; Gottardello, Priscila; Adams, David G. (2007). "Molecular Analysis of Genes in Nostoc punctiforme Involved in Pilus Biogenesis and Plant Infection". Journal of Bacteriology. 189 (12): 4547–4551. doi:10.1128/JB.01927-06. PMC 1913353. PMID 17416648.
- Wilde, Annegret; Mullineaux, Conrad W. (2015). "Motility in cyanobacteria: Polysaccharide tracks and Type IV pilus motors". Molecular Microbiology. 98 (6): 998–1001. doi:10.1111/mmi.13242. PMID 26447922. S2CID 22585994.
- Waterbury, J. B.; Willey, J. M.; Franks, D. G.; Valois, F. W.; Watson, S. W. (1985). "A Cyanobacterium Capable of Swimming Motility". Science. 230 (4721): 74–76. Bibcode:1985Sci...230...74W. doi:10.1126/science.230.4721.74. PMID 17817167. S2CID 29180516.
- Ehlers, Kurt; Oster, George (2012). "On the Mysterious Propulsion of Synechococcus". PLOS ONE. 7 (5): e36081. Bibcode:2012PLoSO...736081E. doi:10.1371/journal.pone.0036081. PMC 3342319. PMID 22567124.
- Miyata, Makoto; Robinson, Robert C.; Uyeda, Taro Q. P.; Fukumori, Yoshihiro; Fukushima, Shun‐Ichi; Haruta, Shin; Homma, Michio; Inaba, Kazuo; Ito, Masahiro; Kaito, Chikara; Kato, Kentaro; Kenri, Tsuyoshi; Kinosita, Yoshiaki; Kojima, Seiji; Minamino, Tohru; Mori, Hiroyuki; Nakamura, Shuichi; Nakane, Daisuke; Nakayama, Koji; Nishiyama, Masayoshi; Shibata, Satoshi; Shimabukuro, Katsuya; Tamakoshi, Masatada; Taoka, Azuma; Tashiro, Yosuke; Tulum, Isil; Wada, Hirofumi; Wakabayashi, Ken‐Ichi (2020). "Tree of motility – A proposed history of motility systems in the tree of life". Genes to Cells. 25 (1): 6–21. doi:10.1111/gtc.12737. PMC 7004002. PMID 31957229.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - R. W. Castenholz (1982). "Motility and taxes". In N. G. Carr; B. A. Whitton (eds.). The biology of cyanobacteria. University of California Press, Berkeley and Los Angeles. pp. 413–439. ISBN 978-0-520-04717-4.
- J. B. Waterbury; J. M. Willey; D. G. Franks; F. W. Valois & S. W. Watson (1985). "A cyanobacterium capable of swimming motility". Science. 230 (4721): 74–76. Bibcode:1985Sci...230...74W. doi:10.1126/science.230.4721.74. PMID 17817167. S2CID 29180516.
- McBride, Mark J. (2001). "Bacterial Gliding Motility: Multiple Mechanisms for Cell Movement over Surfaces". Annual Review of Microbiology. 55: 49–75. doi:10.1146/annurev.micro.55.1.49. PMID 11544349.
- Reichenbach, H. (1981). "Taxonomy of the Gliding Bacteria". Annual Review of Microbiology. 35: 339–364. doi:10.1146/annurev.mi.35.100181.002011. PMID 6794424.
- Hoiczyk, Egbert; Baumeister, Wolfgang (1998). "The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria". Current Biology. 8 (21): 1161–1168. doi:10.1016/S0960-9822(07)00487-3. PMID 9799733. S2CID 14384308.
- Hoiczyk, E. (2000). "Gliding motility in cyanobacteria: Observations and possible explanations". Archives of Microbiology. 174 (1–2): 11–17. doi:10.1007/s002030000187. PMID 10985737. S2CID 9927312.
- Bhaya, D.; Watanabe, N.; Ogawa, T.; Grossman, A. R. (1999). "The role of an alternative sigma factor in motility and pilus formation in the cyanobacterium Synechocystis sp. Strain PCC6803". Proceedings of the National Academy of Sciences. 96 (6): 3188–3193. Bibcode:1999PNAS...96.3188B. doi:10.1073/pnas.96.6.3188. PMC 15917. PMID 10077659.
- Donkor, Victoria A.; Amewowor, Damina H.A.K.; Hã¤Der, Donat-P. (1993). "Effects of tropical solar radiation on the motility of filamentous cyanobacteria". FEMS Microbiology Ecology. 12 (2): 143–147. doi:10.1111/j.1574-6941.1993.tb00026.x.
- Garcia-Pichel, Ferran; Mechling, Margaret; Castenholz, Richard W. (1994). "Diel Migrations of Microorganisms within a Benthic, Hypersaline Mat Community". Applied and Environmental Microbiology. 60 (5): 1500–1511. Bibcode:1994ApEnM..60.1500G. doi:10.1128/aem.60.5.1500-1511.1994. PMC 201509. PMID 16349251.
- Fourã§Ans, Aude; Solã©, Antoni; Diestra, Ella; Ranchou-Peyruse, Anthony; Esteve, Isabel; Caumette, Pierre; Duran, Robert (2006). "Vertical migration of phototrophic bacterial populations in a hypersaline microbial mat from Salins-de-Giraud (Camargue, France)". FEMS Microbiology Ecology. 57 (3): 367–377. doi:10.1111/j.1574-6941.2006.00124.x. PMID 16907751.
- Richardson, Laurie L.; Castenholz, Richard W. (1987). "Diel Vertical Movements of the Cyanobacterium Oscillatoria terebriformis in a Sulfide-Rich Hot Spring Microbial Mat". Applied and Environmental Microbiology. 53 (9): 2142–2150. Bibcode:1987ApEnM..53.2142R. doi:10.1128/aem.53.9.2142-2150.1987. PMC 204072. PMID 16347435.
- Häder, Donat-P. (1987). "EFFECTS OF UV-B IRRADIATION ON PHOTOMOVEMENT IN THE DESMID, Cosmarium cucumis". Photochemistry and Photobiology. 46: 121–126. doi:10.1111/j.1751-1097.1987.tb04745.x. S2CID 97100233.
- Nultsch, Wilhelm; Häder, Donat-P. (1988). "Photomovement in Motile Microorganisms—Ii". Photochemistry and Photobiology. 47 (6): 837–869. doi:10.1111/j.1751-1097.1988.tb01668.x. PMID 3064112. S2CID 26445775.
- Checcucci, G., Sgarbossa, A. and Lenci, F. (2004) "Photomovements of microorganisms: An introduction". CRC Handbook of Organic Photochemistry and Photobiology, 2nd ed., CRC Press.
- Nultsch, Wilhelm (1962) "DER EINFLUSS DES LICHTES AUF DIE BEWEGUNG DER CYANOPHYCEEN: III. Mitteilung: PHOTOPHOBOTAXIS VON PHORMIDIUM UNCINATUM." Planta, 58(6 ): 647–63.
- Nultsch, Wilhelm; Schuchart, Hartwig; Höhl, Marga (1979). "Investigations on the phototactic orientation of Anabaena variabilis". Archives of Microbiology. 122: 85–91. doi:10.1007/BF00408050. S2CID 12242837.
- Riding, R. (2007). "The term stromatolite: towards an essential definition". Lethaia. 32 (4): 321–30. doi:10.1111/j.1502-3931.1999.tb00550.x.
- Baumgartner RJ, et al. (2019). "Nano-porous pyrite and organic matter in 3.5-billion-year-old stromatolites record primordial life" (PDF). Geology. 47 (11): 1039–43. Bibcode:2019Geo....47.1039B. doi:10.1130/G46365.1. S2CID 204258554.
- Lane, Nick (6 February 2010) "First breath: Earth's billion-year struggle for oxygen". New Scientist, pp. 36–39. See accompanying graph as well.
- Corsetti FA, Awramik SM, Pierce D (April 2003). "A complex microbiota from snowball Earth times: microfossils from the Neoproterozoic Kingston Peak Formation, Death Valley, USA". Proceedings of the National Academy of Sciences of the United States of America. 100 (8): 4399–404. Bibcode:2003PNAS..100.4399C. doi:10.1073/pnas.0730560100. PMC 153566. PMID 12682298.
- Gutschick RC, Perry TG (1 November 1959). "Sappington (Kinderhookian) sponges and their environment [Montana]". Journal of Paleontology. 33 (6): 977–85. Retrieved 28 June 2007.
- Riding, Robert. (1991). Calcareous Algae and Stromatolites, p. 32. Springer-Verlag Press.
- Monty, C. L. (1981). Monty, Claude (ed.). "Spongiostromate vs. Porostromate Stromatolites and Oncolites". Phanerozoic Stromatolites. Berlin, Heidelberg: Springer: 1–4. doi:10.1007/978-3-642-67913-1_1. ISBN 978-3-642-67913-1.
- Castellani, Christopher; Maas, Andreas; Eriksson, Mats E.; Haug, Joachim T.; Haug, Carolin; Waloszek, Dieter (2018). "First record of Cyanobacteria in Cambrian Orsten deposits of Sweden". Palaeontology. 61 (6): 855–880. doi:10.1111/pala.12374. S2CID 134049042.
- "How do plants make oxygen? Ask cyanobacteria". Phys.org. Science X. 30 March 2017. Retrieved 26 October 2017.
- Herrero A (2008). Flores E (ed.). The Cyanobacteria: Molecular Biology, Genomics and Evolution (1st ed.). Caister Academic Press. ISBN 978-1-904455-15-8.
- Keeling PJ (2013). "The number, speed, and impact of plastid endosymbioses in eukaryotic evolution". Annual Review of Plant Biology. 64: 583–607. doi:10.1146/annurev-arplant-050312-120144. PMID 23451781. S2CID 207679266.
- Moore KR, Magnabosco C, Momper L, Gold DA, Bosak T, Fournier GP (2019). "An Expanded Ribosomal Phylogeny of Cyanobacteria Supports a Deep Placement of Plastids". Frontiers in Microbiology. 10: 1612. doi:10.3389/fmicb.2019.01612. PMC 6640209. PMID 31354692.
- Howe CJ, Barbrook AC, Nisbet RE, Lockhart PJ, Larkum AW (August 2008). "The origin of plastids". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 363 (1504): 2675–85. doi:10.1098/rstb.2008.0050. PMC 2606771. PMID 18468982.
- Rodríguez-Ezpeleta N, Brinkmann H, Burey SC, Roure B, Burger G, Löffelhardt W, Bohnert HJ, Philippe H, Lang BF (July 2005). "Monophyly of primary photosynthetic eukaryotes: green plants, red algae, and glaucophytes". Current Biology. 15 (14): 1325–30. doi:10.1016/j.cub.2005.06.040. PMID 16051178.
- Adl SM, Simpson AG, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, Heiss A, Hoppenrath M, Lara E, Le Gall L, Lynn DH, McManus H, Mitchell EA, Mozley-Stanridge SE, Parfrey LW, Pawlowski J, Rueckert S, Shadwick L, Shadwick L, Schoch CL, Smirnov A, Spiegel FW (September 2012). "The revised classification of eukaryotes". The Journal of Eukaryotic Microbiology. 59 (5): 429–93. doi:10.1111/j.1550-7408.2012.00644.x. PMC 3483872. PMID 23020233.
- Price DC, Chan CX, Yoon HS, Yang EC, Qiu H, Weber AP, Schwacke R, Gross J, Blouin NA, Lane C, Reyes-Prieto A, Durnford DG, Neilson JA, Lang BF, Burger G, Steiner JM, Löffelhardt W, Meuser JE, Posewitz MC, Ball S, Arias MC, Henrissat B, Coutinho PM, Rensing SA, Symeonidi A, Doddapaneni H, Green BR, Rajah VD, Boore J, Bhattacharya D (February 2012). "Cyanophora paradoxa genome elucidates origin of photosynthesis in algae and plants". Science. 335 (6070): 843–47. Bibcode:2012Sci...335..843P. doi:10.1126/science.1213561. PMID 22344442. S2CID 17190180.
- Ponce-Toledo RI, Deschamps P, López-García P, Zivanovic Y, Benzerara K, Moreira D (February 2017). "An Early-Branching Freshwater Cyanobacterium at the Origin of Plastids". Current Biology. 27 (3): 386–91. doi:10.1016/j.cub.2016.11.056. PMC 5650054. PMID 28132810.
- Schimper, AF (1883). "Über die Entwicklung der Chlorophyllkörner und Farbkörper" [About the development of the chlorophyll grains and stains]. Bot. Zeitung (in German). 41: 105–14, 121–31, 137–46, 153–62. Archived from the original on 19 October 2013.
- Alberts B (2002). Molecular biology of the cell (4. ed.). New York [u.a.]: Garland. ISBN 978-0-8153-4072-0.
- Mereschkowsky C (1905). "Über Natur und Ursprung der Chromatophoren im Pflanzenreiche" [About the nature and origin of chromatophores in the vegetable kingdom]. Biol Centralbl (in German). 25: 593–604.
- Sagan L (March 1967). "On the origin of mitosing cells". Journal of Theoretical Biology. 14 (3): 255–74. Bibcode:1967JThBi..14..225S. doi:10.1016/0022-5193(67)90079-3. PMID 11541392.
- Schwartz RM, Dayhoff MO (January 1978). "Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts". Science. 199 (4327): 395–403. Bibcode:1978Sci...199..395S. doi:10.1126/science.202030. PMID 202030.
- Archibald JM (August 2015). "Genomic perspectives on the birth and spread of plastids". Proceedings of the National Academy of Sciences of the United States of America. 112 (33): 10147–53. Bibcode:2015PNAS..11210147A. doi:10.1073/pnas.1421374112. PMC 4547232. PMID 25902528.
- Blankenship RE (October 2010). "Early evolution of photosynthesis". Plant Physiology. 154 (2): 434–38. doi:10.1104/pp.110.161687. PMC 2949000. PMID 20921158.
- Rockwell NC, Lagarias JC, Bhattacharya D (2014). "Primary endosymbiosis and the evolution of light and oxygen sensing in photosynthetic eukaryotes". Frontiers in Ecology and Evolution. 2 (66). doi:10.3389/fevo.2014.00066. PMC 4343542. PMID 25729749.
- Summarised in Cavalier-Smith T (April 2000). "Membrane heredity and early chloroplast evolution". Trends in Plant Science. 5 (4): 174–82. doi:10.1016/S1360-1385(00)01598-3. PMID 10740299.
- Nowack EC, Melkonian M, Glöckner G (March 2008). "Chromatophore genome sequence of Paulinella sheds light on acquisition of photosynthesis by eukaryotes". Current Biology. 18 (6): 410–18. doi:10.1016/j.cub.2008.02.051. PMID 18356055.
- Keeling PJ (2004). "Diversity and evolutionary history of plastids and their hosts". American Journal of Botany. 91 (10): 1481–1493. doi:10.3732/ajb.91.10.1481. PMID 21652304.
- Archibald JM (January 2009). "The puzzle of plastid evolution". Current Biology. 19 (2): R81–88. doi:10.1016/j.cub.2008.11.067. PMID 19174147.
- Douglas SE (1998). "Plastid evolution: origins, diversity, trends". Curr. Opin. Genet. Dev. 8 (6): 655–61. doi:10.1016/S0959-437X(98)80033-6. PMID 9914199.
- Reyes-Prieto A, Weber AP, Bhattacharya D (2007). "The origin and establishment of the plastid in algae and plants". Annu. Rev. Genet. 41: 147–68. doi:10.1146/annurev.genet.41.110306.130134. PMID 17600460.
- Raven JA, Allen JF (2003). "Genomics and chloroplast evolution: what did cyanobacteria do for plants?". Genome Biol. 4 (3): 209. doi:10.1186/gb-2003-4-3-209. PMC 153454. PMID 12620099.
- Bekker, A.; Holland, H. D.; Wang, P.-L.; Rumble, D.; Stein, H. J.; Hannah, J. L.; Coetzee, L. L.; Beukes, N. J. (2004). "Dating the rise of atmospheric oxygen". Nature. 427 (6970): 117–120. Bibcode:2004Natur.427..117B. doi:10.1038/nature02260. PMID 14712267. S2CID 4329872.
- Kump, L. R.; Junium, C.; Arthur, M. A.; Brasier, A.; Fallick, A.; Melezhik, V.; Lepland, A.; Ccrne, A. E.; Luo, G. (2011). "Isotopic Evidence for Massive Oxidation of Organic Matter Following the Great Oxidation Event". Science. 334 (6063): 1694–1696. Bibcode:2011Sci...334.1694K. doi:10.1126/science.1213999. PMID 22144465. S2CID 27680273.
- Fralick, Philip; Davis, Don W.; Kissin, Stephen A. (2002). "The age of the Gunflint Formation, Ontario, Canada: Single zircon U–Pb age determinations from reworked volcanic ash". Canadian Journal of Earth Sciences. 39 (7): 1085–1091. Bibcode:2002CaJES..39.1085F. doi:10.1139/e02-028.
- Holland, Heinrich D. (2006). "The oxygenation of the atmosphere and oceans". Philosophical Transactions of the Royal Society B: Biological Sciences. 361 (1470): 903–915. doi:10.1098/rstb.2006.1838. PMC 1578726. PMID 16754606.
- Lyons, Timothy W.; Reinhard, Christopher T.; Planavsky, Noah J. (2014). "The rise of oxygen in Earth's early ocean and atmosphere". Nature. 506 (7488): 307–315. Bibcode:2014Natur.506..307L. doi:10.1038/nature13068. PMID 24553238. S2CID 4443958.
- Sahoo, Swapan K.; Planavsky, Noah J.; Kendall, Brian; Wang, Xinqiang; Shi, Xiaoying; Scott, Clint; Anbar, Ariel D.; Lyons, Timothy W.; Jiang, Ganqing (2012). "Ocean oxygenation in the wake of the Marinoan glaciation". Nature. 489 (7417): 546–549. Bibcode:2012Natur.489..546S. doi:10.1038/nature11445. PMID 23018964. S2CID 4391882.
- Och, Lawrence M.; Shields-Zhou, Graham A. (2012). "The Neoproterozoic oxygenation event: Environmental perturbations and biogeochemical cycling". Earth-Science Reviews. 110 (1–4): 26–57. Bibcode:2012ESRv..110...26O. doi:10.1016/j.earscirev.2011.09.004.
- Canfield, D. E.; Poulton, S. W.; Narbonne, G. M. (2007). "Late-Neoproterozoic Deep-Ocean Oxygenation and the Rise of Animal Life". Science. 315 (5808): 92–95. Bibcode:2007Sci...315...92C. doi:10.1126/science.1135013. PMID 17158290. S2CID 24761414.
- Fike, D. A.; Grotzinger, J. P.; Pratt, L. M.; Summons, R. E. (2006). "Oxidation of the Ediacaran Ocean". Nature. 444 (7120): 744–747. Bibcode:2006Natur.444..744F. doi:10.1038/nature05345. PMID 17151665. S2CID 4337003.
- Sánchez-Baracaldo, Patricia; Ridgwell, Andy; Raven, John A. (2014). "A Neoproterozoic Transition in the Marine Nitrogen Cycle". Current Biology. 24 (6): 652–657. doi:10.1016/j.cub.2014.01.041. PMID 24583016. S2CID 16756351.
- Erwin, D. H.; Laflamme, M.; Tweedt, S. M.; Sperling, E. A.; Pisani, D.; Peterson, K. J. (2011). "The Cambrian Conundrum: Early Divergence and Later Ecological Success in the Early History of Animals". Science. 334 (6059): 1091–1097. Bibcode:2011Sci...334.1091E. doi:10.1126/science.1206375. PMID 22116879. S2CID 7737847.
- Poulton, S. W.; Canfield, D. E. (2011). "Ferruginous Conditions: A Dominant Feature of the Ocean through Earth's History". Elements. 7 (2): 107–112. doi:10.2113/gselements.7.2.107.
- Anbar, A. D.; Knoll, A. H. (2002). "Proterozoic Ocean Chemistry and Evolution: A Bioinorganic Bridge?". Science. 297 (5584): 1137–1142. Bibcode:2002Sci...297.1137A. doi:10.1126/science.1069651. PMID 12183619. S2CID 5578019.
- Orkwiszewski KG, Kaney AR (June 1974). "Genetic transformation of the blue-green bacterium, Anacystis nidulans". Arch Mikrobiol. 98 (1): 31–37. doi:10.1007/BF00425265. PMID 4209657. S2CID 5635245.
- Stevens SE, Porter RD (October 1980). "Transformation in Agmenellum quadruplicatum". Proc. Natl. Acad. Sci. U.S.A. 77 (10): 6052–56. Bibcode:1980PNAS...77.6052S. doi:10.1073/pnas.77.10.6052. PMC 350211. PMID 16592896.
- Grigorieva G, Shestakov S (1982). "Transformation in the cyanobacterium Synechocystis sp. 6803". FEMS Microbiology Letters. 13 (4): 367–70. doi:10.1111/j.1574-6968.1982.tb08289.x.
- Bernstein H, Bernstein C, Michod RE (January 2018). "Sex in microbial pathogens". Infect. Genet. Evol. 57: 8–25. doi:10.1016/j.meegid.2017.10.024. PMID 29111273.
- Cassier-Chauvat C, Veaudor T, Chauvat F (2016). "Comparative Genomics of DNA Recombination and Repair in Cyanobacteria: Biotechnological Implications". Front Microbiol. 7: 1809. doi:10.3389/fmicb.2016.01809. PMC 5101192. PMID 27881980.
- "The LTP". Retrieved 23 February 2021.
- "LTP_all tree in newick format". Retrieved 23 February 2021.
- "LTP_12_2021 Release Notes" (PDF). Retrieved 23 February 2021.
- "GTDB release 07-RS207". Genome Taxonomy Database. Retrieved 20 June 2022.
- "ar53_r207.sp_label". Genome Taxonomy Database. Retrieved 20 June 2022.
- "Taxon History". Genome Taxonomy Database. Retrieved 20 June 2022.
- Von Nägeli C (1857). Caspary R (ed.). "Bericht über die Verhandlungen der 33. Versammlung deutscher Naturforscher und Ärzte, gehalten in Bonn von 18 bis 24 September 1857" [Report on the Proceedings of the 33rd Meeting of German Natural Scientists and Physicians, held in Bonn, 18 to 24 September 1857]. Botanische Zeitung. 15: 749–776.
- Haeckel E (1867). Generelle Morphologie der Organismen. Reimer, Berlin.
- Chatton É (1925). "Pansporella perplexa: amœbien à spores protégées parasite des daphnies réflexions sur la biologie et la phylogénie des protozoaires" [Pansporella perplexa: amoebian with protected spores parasite of daphnia reflections on the biology and phylogeny of protozoa]. Ann. Sci. Nat. Zool. 10 (in French). VII: 1–84.
- Gugger MF, Hoffmann L (March 2004). "Polyphyly of true branching cyanobacteria (Stigonematales)". International Journal of Systematic and Evolutionary Microbiology. 54 (Pt 2): 349–57. doi:10.1099/ijs.0.02744-0. PMID 15023942.
- Howard-Azzeh M, Shamseer L, Schellhorn HE, Gupta RS (November 2014). "Phylogenetic analysis and molecular signatures defining a monophyletic clade of heterocystous cyanobacteria and identifying its closest relatives". Photosynthesis Research. 122 (2): 171–85. doi:10.1007/s11120-014-0020-x. PMID 24917519. S2CID 17745718.
- Komárek J, Kaštovský J, Mareš J, Johansen JR (2014). "Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach" (PDF). Preslia. 86: 295–335.
- Pringsheim EG (1963). Farblose Algen: Ein Beitrag zur Evolutionsforschung. Gustav Fischer Verlag.
- J.P. Euzéby. ""Cyanobacteria"". List of Prokaryotic names with Standing in Nomenclature (LPSN). Retrieved 22 January 2022.
- Sayers; et al. "Cyanobacteria". National Center for Biotechnology Information (NCBI) taxonomy database. Retrieved 20 March 2021.
- Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S (June 1996). "Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions". DNA Research. 3 (3): 109–36. doi:10.1093/dnares/3.3.109. PMID 8905231.
- Tabei Y, Okada K, Tsuzuki M (April 2007). "Sll1330 controls the expression of glycolytic genes in Synechocystis sp. PCC 6803". Biochemical and Biophysical Research Communications. 355 (4): 1045–50. doi:10.1016/j.bbrc.2007.02.065. PMID 17331473.
- Rocap G, Larimer FW, Lamerdin J, Malfatti S, Chain P, Ahlgren NA, Arellano A, Coleman M, Hauser L, Hess WR, Johnson ZI, Land M, Lindell D, Post AF, Regala W, Shah M, Shaw SL, Steglich C, Sullivan MB, Ting CS, Tolonen A, Webb EA, Zinser ER, Chisholm SW (August 2003). "Genome divergence in two Prochlorococcus ecotypes reflects oceanic niche differentiation". Nature. 424 (6952): 1042–47. Bibcode:2003Natur.424.1042R. doi:10.1038/nature01947. PMID 12917642. S2CID 4344597.
- Dufresne A, Salanoubat M, Partensky F, Artiguenave F, Axmann IM, Barbe V, Duprat S, Galperin MY, Koonin EV, Le Gall F, Makarova KS, Ostrowski M, Oztas S, Robert C, Rogozin IB, Scanlan DJ, Tandeau de Marsac N, Weissenbach J, Wincker P, Wolf YI, Hess WR (August 2003). "Genome sequence of the cyanobacterium Prochlorococcus marinus SS120, a nearly minimal oxyphototrophic genome". Proceedings of the National Academy of Sciences of the United States of America. 100 (17): 10020–25. Bibcode:2003PNAS..10010020D. doi:10.1073/pnas.1733211100. PMC 187748. PMID 12917486.
- Meeks JC, Elhai J, Thiel T, Potts M, Larimer F, Lamerdin J, Predki P, Atlas R (2001). "An overview of the genome of Nostoc punctiforme, a multicellular, symbiotic cyanobacterium". Photosynthesis Research. 70 (1): 85–106. doi:10.1023/A:1013840025518. PMID 16228364. S2CID 8752382.
- Herdman M, Janvier M, Rippka R, Stanier RY (1979). "Genome Size of Cyanobacteria". Journal of General Microbiology. 111: 73–85. doi:10.1099/00221287-111-1-73.
- Quintana N, Van der Kooy F, Van de Rhee MD, Voshol GP, Verpoorte R (August 2011). "Renewable energy from Cyanobacteria: energy production optimization by metabolic pathway engineering". Applied Microbiology and Biotechnology. 91 (3): 471–90. doi:10.1007/s00253-011-3394-0. PMC 3136707. PMID 21691792.
- Chen, Xiaolong; Lawrence, Joshua M.; Wey, Laura T.; Schertel, Lukas; Jing, Qingshen; Vignolini, Silvia; Howe, Christopher J.; Kar-Narayan, Sohini; Zhang, Jenny Z. (7 March 2022). "3D-printed hierarchical pillar array electrodes for high-performance semi-artificial photosynthesis". Nature Materials. 21 (7): 811–818. Bibcode:2022NatMa..21..811C. doi:10.1038/s41563-022-01205-5. ISSN 1476-4660. PMID 35256790. S2CID 247255146.
- "Blue green bacteria may help generate 'green' electricity", The Hindu, 21 June 2010
- "Joule wins key patent for GMO cyanobacteria that create fuels from sunlight, CO2 and water : Biofuels Digest". 14 September 2010. Retrieved 10 August 2022.
- Deng, Ming-De; Coleman, John R. (1 February 1999). "Ethanol Synthesis by Genetic Engineering in Cyanobacteria". Applied and Environmental Microbiology. 65 (2): 523–528. Bibcode:1999ApEnM..65..523D. doi:10.1128/AEM.65.2.523-528.1999. PMC 91056. PMID 9925577.
- Liang, Feiyan; Lindberg, Pia; Lindblad, Peter (20 November 2018). "Engineering photoautotrophic carbon fixation for enhanced growth and productivity". Sustainable Energy & Fuels. 2 (12): 2583–2600. doi:10.1039/C8SE00281A. Retrieved 15 February 2022 – via pubs.rsc.org.
- Roussou, S., Albergati, A., Liang, F., & Lindblad, P. (2021). Engineered cyanobacteria with additional overexpression of selected Calvin-Benson-Bassham enzymes show further increased ethanol production. Metabolic Engineering Communications, 12, e00161.
- Choi H, Mascuch SJ, Villa FA, Byrum T, Teasdale ME, Smith JE, et al. (May 2012). "Honaucins A-C, potent inhibitors of inflammation and bacterial quorum sensing: synthetic derivatives and structure-activity relationships". Chemistry & Biology. 19 (5): 589–98. doi:10.1016/j.chembiol.2012.03.014. PMC 3361693. PMID 22633410.
- Frischkorn, Kyle (19 June 2017). "Need to Fix a Heart Attack? Try Photosynthesis". Smithsonian. Retrieved 20 May 2021.
- Jeong, Yujin; Cho, Sang-Hyeok; Lee, Hookeun; Choi, Hyung-Kyoon; Kim, Dong-Myung; Lee, Choul-Gyun; Cho, Suhyung; Cho, Byung-Kwan (December 2020). "Current Status and Future Strategies to Increase Secondary Metabolite Production from Cyanobacteria". Microorganisms. 8 (12): 1849. doi:10.3390/microorganisms8121849. PMC 7761380. PMID 33255283.
- Newsome, Andrew G.; Culver, Catherine A.; van Breemen, Richard B. (16 July 2014). "Nature's Palette: The Search for Natural Blue Colorants". Journal of Agricultural and Food Chemistry. 62 (28): 6498–6511. doi:10.1021/jf501419q. ISSN 0021-8561. PMID 24930897.
- Verseux C, Baqué M, Lehto K, de Vera JP, Rothschild LJ, Billi D (2016). "Sustainable life support on Mars – the potential roles of cyanobacteria". International Journal of Astrobiology. 15 (1): 65–92. Bibcode:2016IJAsB..15...65V. doi:10.1017/S147355041500021X.
- Spolaore P, Joannis-Cassan C, Duran E, Isambert A (February 2006). "Commercial applications of microalgae". Journal of Bioscience and Bioengineering. 101 (2): 87–96. doi:10.1263/jbb.101.87. PMID 16569602. S2CID 16896655.
- Christaki E, Florou-Paneri P, Bonos E (December 2011). "Microalgae: a novel ingredient in nutrition". International Journal of Food Sciences and Nutrition. 62 (8): 794–99. doi:10.3109/09637486.2011.582460. PMID 21574818. S2CID 45956239.
- Ku CS, Pham TX, Park Y, Kim B, Shin MS, Kang I, Lee J (April 2013). "Edible blue-green algae reduce the production of pro-inflammatory cytokines by inhibiting NF-κB pathway in macrophages and splenocytes". Biochimica et Biophysica Acta (BBA) - General Subjects. 1830 (4): 2981–88. doi:10.1016/j.bbagen.2013.01.018. PMC 3594481. PMID 23357040.
- Mišurcová L, Škrovánková S, Samek D, Ambrožová J, Machů L (2012). "Health benefits of algal polysaccharides in human nutrition". Advances in Food and Nutrition Research Volume 66. Advances in Food and Nutrition Research. Vol. 66. pp. 75–145. doi:10.1016/B978-0-12-394597-6.00003-3. ISBN 978-0-12-394597-6. PMID 22909979.
- Thébault L, Lesne J, Boutin JP (1995). "[Cyanobacteria, their toxins and health risks]". Médecine Tropicale. 55 (4): 375–80. PMID 8830224.
- "Blue-Green Algae (Cyanobacteria) and Their Toxins - Drinking Water". 30 May 2008. Archived from the original on 30 May 2008. Retrieved 10 August 2022.
{{cite web}}: CS1 maint: bot: original URL status unknown (link) - "Harmful Bloom in Lake Atitlán, Guatemala". earthobservatory.nasa.gov. 3 December 2009. Retrieved 10 August 2022. from NASA Earth Observatory,
- Weinrich C, Miller TR (2014). "Freshwater harmful algal blooms: toxins and children's health". Current Problems in Pediatric and Adolescent Health Care. 44 (1): 2–24. doi:10.1016/j.cppeds.2013.10.007. PMID 24439026.
- "Professor Linda Lawton". rgu-repository.worktribe.com. Retrieved 25 June 2021.
- Caller TA, Doolin JW, Haney JF, Murby AJ, West KG, Farrar HE, Ball A, Harris BT, Stommel EW (2009). "A cluster of amyotrophic lateral sclerosis in New Hampshire: a possible role for toxic cyanobacteria blooms". Amyotrophic Lateral Sclerosis. 10 (Suppl 2): 101–08. doi:10.3109/17482960903278485. PMID 19929741. S2CID 35250897.
- Cox PA, Richer R, Metcalf JS, Banack SA, Codd GA, Bradley WG (2009). "Cyanobacteria and BMAA exposure from desert dust: a possible link to sporadic ALS among Gulf War veterans". Amyotrophic Lateral Sclerosis. 10 (Suppl 2): 109–17. doi:10.3109/17482960903286066. PMID 19929742. S2CID 1748627.
- Main D (2006). "Toxic Algae Blooms" (PDF). Veterinary Pathologist, South Perth. agric.wa.gov.au. Archived from the original (PDF) on 1 December 2014. Retrieved 18 November 2014.
- May V, Baker H (1978). "Reduction of toxic algae in farm dams by ferric alum". Techn. Bull. 19: 1–16.
- Paerl, H. W.; Huisman, J. (2008). "CLIMATE: Blooms Like It Hot". Science. 320 (5872): 57–58. doi:10.1126/science.1155398. PMID 18388279. S2CID 142881074.
- Sitoki, L.; Kurmayer, R.; Rott, E. (2012). "Spatial variation of phytoplankton composition, biovolume, and resulting microcystin concentrations in the Nyanza Gulf (Lake Victoria, Kenya)". Hydrobiologia. 691 (1): 109–122. doi:10.1007/s10750-012-1062-8. PMC 3968937. PMID 24683268.
- Cox, Paul Alan; Brand, Larry E.; Murch, Susan J.; Tymm, Fiona J. M.; Powell, James T.; Banack, Sandra Anne; Metcalf, James S. (2018). "Public health responses to toxic cyanobacterial blooms: Perspectives from the 2016 Florida event". Water Policy. 20 (5): 919–932. doi:10.2166/wp.2018.012. S2CID 88502303.
- Cavicchioli, Ricardo; Ripple, William J.; Timmis, Kenneth N.; Azam, Farooq; Bakken, Lars R.; Baylis, Matthew; Behrenfeld, Michael J.; Boetius, Antje; Boyd, Philip W.; Classen, Aimée T.; Crowther, Thomas W.; Danovaro, Roberto; Foreman, Christine M.; Huisman, Jef; Hutchins, David A.; Jansson, Janet K.; Karl, David M.; Koskella, Britt; Mark Welch, David B.; Martiny, Jennifer B. H.; Moran, Mary Ann; Orphan, Victoria J.; Reay, David S.; Remais, Justin V.; Rich, Virginia I.; Singh, Brajesh K.; Stein, Lisa Y.; Stewart, Frank J.; Sullivan, Matthew B.; et al. (2019). "Scientists' warning to humanity: Microorganisms and climate change". Nature Reviews Microbiology. 17 (9): 569–586. doi:10.1038/s41579-019-0222-5. PMC 7136171. PMID 31213707.
 Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Material was copied from this source, which is available under a Creative Commons Attribution 4.0 International License. - Visser, Petra M.; Verspagen, Jolanda M.H.; Sandrini, Giovanni; Stal, Lucas J.; Matthijs, Hans C.P.; Davis, Timothy W.; Paerl, Hans W.; Huisman, Jef (2016). "How rising CO2 and global warming may stimulate harmful cyanobacterial blooms". Harmful Algae. 54: 145–159. doi:10.1016/j.hal.2015.12.006. PMID 28073473.
- Walsby, Anthony E.; Hayes, Paul K.; Boje, Rolf; Stal, Lucas J. (1997). "The selective advantage of buoyancy provided by gas vesicles for planktonic cyanobacteria in the Baltic Sea". New Phytologist. 136 (3): 407–417. doi:10.1046/j.1469-8137.1997.00754.x. PMID 33863010.
- Jöhnk, Klaus D.; Huisman, JEF; Sharples, Jonathan; Sommeijer, BEN; Visser, Petra M.; Stroom, Jasper M. (2008). "Summer heatwaves promote blooms of harmful cyanobacteria". Global Change Biology. 14 (3): 495–512. Bibcode:2008GCBio..14..495J. doi:10.1111/j.1365-2486.2007.01510.x. S2CID 54079634.
- Lehman, P.W.; Kurobe, T.; Lesmeister, S.; Baxa, D.; Tung, A.; Teh, S.J. (2017). "Impacts of the 2014 severe drought on the Microcystis bloom in San Francisco Estuary". Harmful Algae. 63: 94–108. doi:10.1016/j.hal.2017.01.011. PMID 28366405.
- Sandrini, Giovanni; Ji, Xing; Verspagen, Jolanda M. H.; Tann, Robert P.; Slot, Pieter C.; Luimstra, Veerle M.; Schuurmans, J. Merijn; Matthijs, Hans C. P.; Huisman, Jef (2016). "Rapid adaptation of harmful cyanobacteria to rising CO2". Proceedings of the National Academy of Sciences. 113 (33): 9315–9320. Bibcode:2016PNAS..113.9315S. doi:10.1073/pnas.1602435113. PMC 4995953. PMID 27482094.
- Attribution
![]() This article incorporates text available under the CC BY 2.5 license.
This article incorporates text available under the CC BY 2.5 license.
Further reading
- Cribbs G (1997). Nature's Superfood: the Blue-Green Algae Revolution (first ed.). Newleaf. ISBN 978-0-7522-0569-4.
- Savage M (1994). The Millennial Project: Colonizing the Galaxy in Eight Easy Steps. Little Brown & Co. ISBN 978-0-316-77163-4.
- Fogg GE, Stewart WD, Fay P, Walsby AE (1973). The Blue-green Algae. London and New York: Academic Press. ISBN 978-0-12-261650-1.
- "Architects of the earth's atmosphere", Introduction to the Cyanobacteria, University of California, Berkeley, 3 February 2006.
- Whitton BA (2002). "Phylum Cyanophyta (Cyanobacteria)". The Freshwater Algal Flora of the British Isles. Cambridge: Cambridge University Press. ISBN 978-0-521-77051-4.
- Pentecost A, Franke U (2010). "Photosynthesis and calcification of the stromatolitic freshwater cyanobacterium Rivularia". Eur. J. Phycol. 45 (4): 345–53. doi:10.1080/09670262.2010.492914.
- Whitton BA, Potts M, eds. (2000). The Ecology of Cyanobacteria: their Diversity in Time and Space. Springer. ISBN 978-0-7923-4735-4.
- "From Micro-Algae to Blue Oil". ParisTech Review. December 2011. Archived from the original on 17 April 2016. Retrieved 2 March 2012.