Bunyavirales
| Bunyavirales | |
|---|---|
 | |
| Crimean-Congo hemorrhagic fever virus (CCHFV) virion and replication cycle | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Negarnaviricota |
| Subphylum: | Polyploviricotina |
| Class: | Ellioviricetes |
| Order: | Bunyavirales |
| Families[1] | |
| |
Bunyavirales is an order of segmented negative-strand RNA viruses with mainly tripartite genomes. Member viruses infect arthropods, plants, protozoans, and vertebrates.[2] It is the only order in the class Ellioviricetes.[1] The name Bunyavirales derives from Bunyamwera,[3] where the original type species Bunyamwera orthobunyavirus was first discovered.[4] Ellioviricetes is named in honor of late virologist Richard M. Elliott for his early work on bunyaviruses.[5]
Bunyaviruses belong to the fifth group of the Baltimore classification system, which includes viruses with a negative-sense, single-stranded RNA genome. They have an enveloped, spherical virion. Though generally found in arthropods or rodents, certain viruses in this order occasionally infect humans. Some of them also infect plants.[6] In addition, there is a group of bunyaviruses whose replication is restricted to arthropods and is known as insect-specific bunyaviruses.[7]
A majority of bunyaviruses are vector-borne. With the exception of Hantaviruses and Arenaviruses, all viruses in the Bunyavirales order are transmitted by arthropods (mosquitos, tick, or sandfly). Hantaviruses are transmitted through contact with rodent feces. Incidence of infection is closely linked to vector activity, for example, mosquito-borne viruses are more common in the summer.[6]
Human infections with certain members of Bunyavirales, such as Crimean-Congo hemorrhagic fever orthonairovirus, are associated with high levels of morbidity and mortality, consequently handling of these viruses is done in biosafety level 4 laboratories. They are also the cause of severe fever with thrombocytopenia syndrome.[8]
Hantaviruses are another medically important member of the order Bunyvirales. They are found worldwide, and are relatively common in Korea, Scandinavia (including Finland), Russia, western North America and parts of South America. Hantavirus infections are associated with high fever, lung edema, and pulmonary failure. The mortality rate varies significantly depending on the form, being up to 50% in New World hantaviruses (the Americas), up to 15% in Old World hantaviruses (Asia and Europe), and as little as 0.1% in Puumala virus (mostly Scandinavia).[9]
Virology
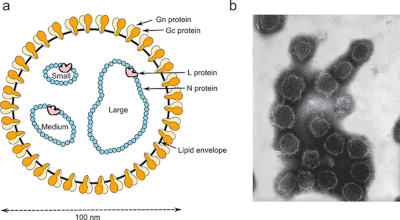
Structure
Bunyavirus morphology is somewhat similar to that of the Paramyxoviridae family; Bunyavirales form enveloped, spherical virions with diameters of 80–120 nm. These viruses contain no matrix proteins.[10]
Genome
Bunyaviruses have bi- or tripartite genomes consisting of a large (L) and small(s), or large (L), medium (M), and small (S) RNA segment. These RNA segments are single-stranded, and exist in a helical formation within the virion. Besides, they exhibit a pseudo-circular structure due to each segment's complementary ends. The L segment encodes the RNA-dependent RNA polymerase, necessary for viral RNA replication and mRNA synthesis. The M segment encodes the viral glycoproteins, which project from the viral surface and aid the virus in attaching to and entering the host cell. The S segment encodes the nucleocapsid protein (N).[11]
Most bunyaviruses have a negative-sense L and M segment. The S segment of the genus Phlebovirus,[12] and both M and S segment of the genus Tospovirus are ambisense.[13] Ambisense means that some of the genes on the RNA strand are negative sense and others are positive sense. The ambisense S segment codes for the viral nucleoprotein (N) in the negative sense and a nonstructural protein (NSs) in the positive sense. The ambisense M segment codes for glycoprotein (GP) in the negative sense and a nonstructural protein (NSm) in the positive sense.[13]
The total genome size ranges from 10.5 to 22.7 kbp.[14]
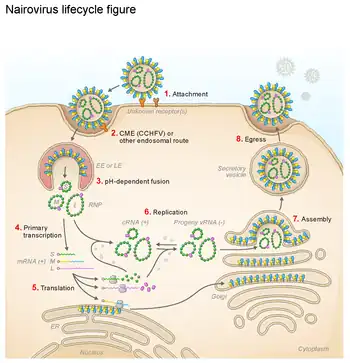
Life cycle
The ambisense genome requires two rounds of transcription to be carried out. First, the negative-sense RNA is transcribed to produce mRNA and a full-length replicative intermediate. [15][16][17]
From this intermediate, a subgenomic mRNA encoding the small segment nonstructural protein is produced while the polymerase produced following the first round of transcription begins to replicate the full-length RNA to produce viral genomes.[15][16][17]
Bunyaviruses replicate in the cytoplasm, while the viral proteins transit through the ER and Golgi apparatus. Mature virions bud from the Golgi apparatus into vesicles which are transported to the cell surface.[15][16][17]
Transmission
Bunyaviruses infect arthropods, plants, protozoans, and vertebrates.[2]
Plants can host bunyaviruses from the families Tospoviridae and Fimoviridae (e.g. tomato, pigeonpea, melon, wheat, raspberry, redbud, and rose). Members of some families are insect-specific, for example the phasmavirids, first isolated from phantom midges,[18] and since identified in diverse insects including moths, wasps and bees, and other true flies.
Taxonomy
There are 477 virus species recognised in this order.[1] The phylogenetic tree diagram provides a full list of member species and the hosts which they infect.[2] The order is organized into the following 12 families:[1]
- Arenaviridae
- Cruliviridae
- Fimoviridae
- Hantaviridae
- Leishbunyaviridae
- Mypoviridae
- Nairoviridae
- Peribunyaviridae
- Phasmaviridae
- Phenuiviridae
- Tospoviridae
- Wupedeviridae
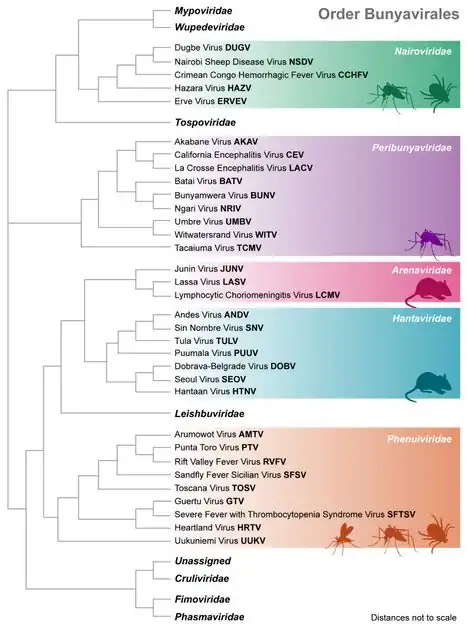 Bunyavirales Phylogenetic tree (bracket representation)
Bunyavirales Phylogenetic tree (bracket representation)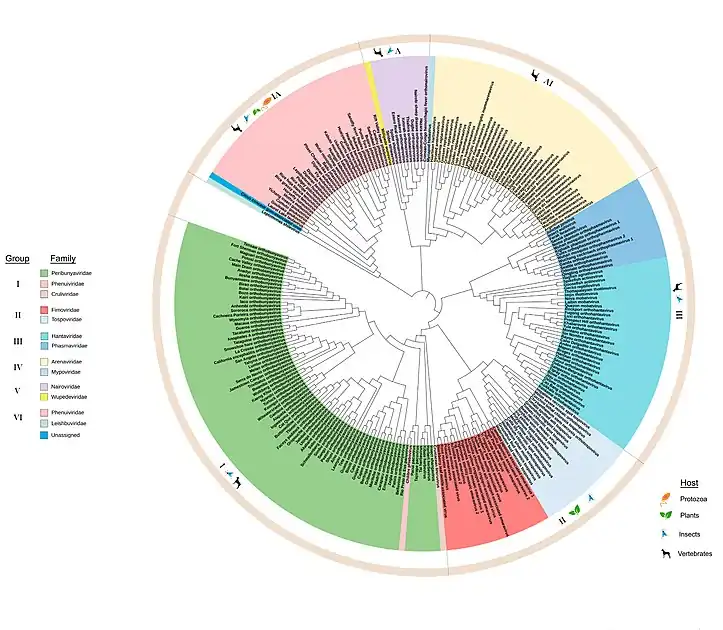 Phylogenetic tree of Bunyavirales ( Maximum likelihood phylogenetic tree of amino acid sequences of RNA-dependent RNA polymerase . The virus families are color-coded and the hosts for viruses within each group are indicated in the outermost circle. )
Phylogenetic tree of Bunyavirales ( Maximum likelihood phylogenetic tree of amino acid sequences of RNA-dependent RNA polymerase . The virus families are color-coded and the hosts for viruses within each group are indicated in the outermost circle. )
Diseases in humans
Bunyaviruses that cause disease in humans include:
- California encephalitis virus, Jamestown Canyon virus, La Crosse encephalitis virus, Oropouche orthobunyavirus (vector: mosquitoes; family: Peribunyaviridae);[19][20]
- Hantavirus reservoir: small mammals or rodents (vector: aerosolized excreta from these mammals; family: Hantaviridae);[21]
- Crimean–Congo hemorrhagic fever reservoir and (vector: ticks; amplifying hosts and vector: small mammals, domestic mammals; family: Nairoviridae);[22]
- Rift Valley fever (reservoir: bats; vector: mosquitoes; amplifying hosts: small and domestic mammals; family: Phenuiviridae);[22]
- Bwamba Fever (reservoir: monkeys; vector: mosquitoes; amplifying hosts: donkeys; family: Peribunyaviridae);[20]
- Severe fever with thrombocytopenia syndrome (vector: ticks);[20]
- Lassa fever (reservoir: rodents; vector: aerosolized excreta from these mammals; family: Arenaviridae).[20]
Bunyaviruses have segmented genomes, making them capable of rapid recombination and increasing the risk of outbreak.[23] The bunyavirus that causes severe fever with thrombocytopenia syndrome can undergo recombination both by reassortment of genome segments and by intragenic homologous recombination.[24][25] Bunyaviridae are transmitted by hematophagous arthropods including mosquitoes, midges, flies, and ticks. The viral incubation period is about 48 hours. Symptomatic infection typically causes non-specific flu-like symptoms with fever lasting for about three days. Because of their non-specific symptoms, Bunyavirus infections are frequently mistaken for other illnesses. For example, Bwamba fever is often mistaken for malaria.[26]
 Male diagnosed with Crimean–Congo hemorrhagic fever
Male diagnosed with Crimean–Congo hemorrhagic fever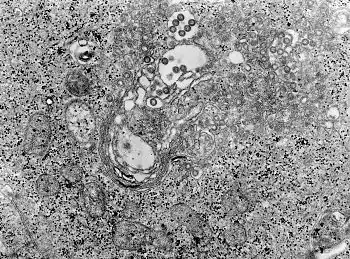 TEM micrograph of tissue infected with Rift Valley fever virus
TEM micrograph of tissue infected with Rift Valley fever virus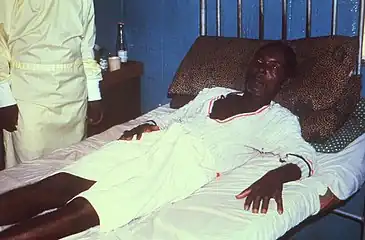 Male with Lassa fever in 1977 lying in a hospital bed in Segbwema, Sierra Leone. Note his dazed, somnolent facial expression due to his weakness, and exhaustion.
Male with Lassa fever in 1977 lying in a hospital bed in Segbwema, Sierra Leone. Note his dazed, somnolent facial expression due to his weakness, and exhaustion. A chest x-ray showing the early stages of bilateral pulmonary effusion due to hantavirus pulmonary syndrome (HPS). HPS begins with interstitial pulmonary edema, progressing to alveolar edema with severe bilateral involvement. Pleural effusions are common.
A chest x-ray showing the early stages of bilateral pulmonary effusion due to hantavirus pulmonary syndrome (HPS). HPS begins with interstitial pulmonary edema, progressing to alveolar edema with severe bilateral involvement. Pleural effusions are common.
Prevention
Prevention depends on the reservoir, amplifying hosts and how the viruses are transmitted, i.e. the vector, whether ticks or mosquitoes and which animals are involved. Preventive measures include general hygiene, limiting contact with vector saliva, urine, feces, or bedding. Currently there is no licensed vaccine for bunyaviruses.[22][27] As a precaution Hantavirus research are conducted in BSL-3 (lab [28]), Rift Valley Fever virus research is conducted in BSL-3 (lab[28]), Congo-Crimean Hemorrhagic Fever virus research is conducted in BSL-4 (lab [28]).
History
In terms of the history of Bunyavirales, we find the following are significant events in its discovery, as well as its epidemiology (or outbreaks), from 1940 to today:
- 1940s: Crimean–Congo hemorrhagic fever is discovered in Crimea[29]
- 1951: 3,000 cases of Hantavirus were reported in South Korea in 1951, a time when UN forces were fighting during the Korean War[30]
- 1956: Cache Valley virus isolated in Culiseta inornata mosquitoes in Utah[31]
- 1960: La Crosse virus was first recognized in a fatal case of encephalitis in La Crosse, Wisconsin[32]
- 1977: Rift Valley Fever virus caused approximately 200,000 cases and 598 deaths in Egypt[33]
- 2017: Bunyavirales order is created[34]
References
- 1 2 3 4 "Virus Taxonomy: 2020 Release". International Committee on Taxonomy of Viruses (ICTV). March 2021. Archived from the original on 20 March 2020. Retrieved 19 May 2021.
- 1 2 3 Herath, Venura; Romay, Gustavo; Urrutia, Cesar D.; Verchot, Jeanmarie (September 2020). "Family Level Phylogenies Reveal Relationships of Plant Viruses within the Order Bunyavirales". Viruses. 12 (9): 1010. doi:10.3390/v12091010. PMC 7551631. PMID 32927652.
- ↑ "ICTV 9th Report (2011) Bunyaviridae". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 11 December 2018. Retrieved 31 January 2019.
Bunya: from Bunyamwera, place in Uganda, where type virus was isolated.
- ↑ Smithburn, K. C.; Haddow, A. J.; Mahaffy, A. F. (March 1946). "A Neurotropic Virus Isolated from Aedes Mosquitoes Caught in the Semliki Forest". The American Journal of Tropical Medicine and Hygiene. s1-26 (2): 189–208. doi:10.4269/ajtmh.1946.s1-26.189. ISSN 1476-1645. OCLC 677158400. PMID 21020339.
- ↑ Wolf, Yuri; Krupovic, Mart; Zhang, Yong Zhen; Maes, Piet; Dolja, Valerian; Koonin, Eugene V.; Kuhn, Jens H. "Megataxonomy of negative-sense RNA viruses". International Committee on Taxonomy of Viruses (ICTV). Archived from the original (docx) on 13 January 2019. Retrieved 12 January 2019.
- 1 2 Plyusnin, A; Elliott, RM, eds. (2011). Bunyaviridae: Molecular and Cellular Biology. Caister Academic Press. ISBN 978-1-904455-90-5.
- ↑ Elrefaey, Ahmed ME; Abdelnabi, Rana; Rosales Rosas, Ana Lucia; Wang, Lanjiao; Basu, Sanjay; Delang, Leen (September 2020). "Understanding the Mechanisms Underlying Host Restriction of Insect-Specific Viruses". Viruses. 12 (9): 964. doi:10.3390/v12090964. PMC 7552076. PMID 32878245.
- ↑ Yu XJ, Liang MF, Zhang SY, et al. (April 2011). "Fever with thrombocytopenia associated with a novel bunyavirus in China". N. Engl. J. Med. 364 (16): 1523–32. doi:10.1056/NEJMoa1010095. PMC 3113718. PMID 21410387.
- ↑ Walter Muranyi; Udo Bahr; Martin Zeier; Fokko J. van der Woude (2005). "Hantavirus Infection". Journal of the American Society of Nephrology. 16 (12): 3669–3679. doi:10.1681/ASN.2005050561. PMID 16267154.
- ↑ "Bunyaviridae - Negative Sense RNA Viruses - Negative Sense RNA Viruses (2011)". International Committee on Taxonomy of Viruses (ICTV). Archived from the original on 2018-12-11. Retrieved 2020-09-08.
- ↑ Ariza, A.; Tanner, S. J.; Walter, C. T.; Dent, K. C.; Shepherd, D. A.; Wu, W.; Matthews, S. V.; Hiscox, J. A.; Green, T. J. (2013-06-01). "Nucleocapsid protein structures from orthobunyaviruses reveal insight into ribonucleoprotein architecture and RNA polymerization". Nucleic Acids Research. 41 (11): 5912–5926. doi:10.1093/nar/gkt268. ISSN 0305-1048. PMC 3675483. PMID 23595147.
- ↑ Elliott, Richard M; Brennan, Benjamin (April 2014). "Emerging phleboviruses". Current Opinion in Virology. 5 (100): 50–57. doi:10.1016/j.coviro.2014.01.011. PMC 4031632. PMID 24607799.
- 1 2 Lima, R. N.; De Oliveira, A. S.; Leastro, M. O.; Blawid, R.; Nagata, T.; Resende, R. O.; Melo, F. L. (7 July 2016). "The complete genome of the tospovirus Zucchini lethal chlorosis virus". Virology Journal. 13 (1): 123. doi:10.1186/s12985-016-0577-4. PMC 4936248. PMID 27388209.
- ↑ "00.011. Bunyaviridae". ICTVdB—The Universal Virus Database, version 4. 2006. Archived from the original on 2008-12-10. Retrieved 2009-01-01.
- 1 2 3 Sun, Yeping; Li, Jing; Gao, George F.; Tien, Po; Liu, Wenjun (3 September 2018). "Bunyavirales ribonucleoproteins: the viral replication and transcription machinery". Critical Reviews in Microbiology. 44 (5): 522–540. doi:10.1080/1040841X.2018.1446901. ISSN 1040-841X. Archived from the original on 20 March 2022. Retrieved 25 January 2023.
- 1 2 3 Ren, Fuli; Shen, Shu; Wang, Qiongya; Wei, Gang; Huang, Chaolin; Wang, Hualin; Ning, Yun-Jia; Zhang, Ding-Yu; Deng, Fei (2021). "Recent Advances in Bunyavirus Reverse Genetics Research: Systems Development, Applications, and Future Perspectives". Frontiers in Microbiology. 12. doi:10.3389/fmicb.2021.771934/full. ISSN 1664-302X. Archived from the original on 9 July 2022. Retrieved 28 January 2023.
- 1 2 3 Papageorgiou, Nicolas; Spiliopoulou, Maria; Nguyen, Thi-Hong Van; Vaitsopoulou, Afroditi; Laban, Elsie Yekwa; Alvarez, Karine; Margiolaki, Irene; Canard, Bruno; Ferron, François (17 July 2020). "Brothers in Arms: Structure, Assembly and Function of Arenaviridae Nucleoprotein". Viruses. 12 (7): 772. doi:10.3390/v12070772. Archived from the original on 2 February 2023. Retrieved 31 January 2023.
- ↑ Ballinger, MJ; Bruenn, JA; Hay, J; Czechowski, D; Taylor, DJ (2014). "Discovery and evolution of bunyavirids in arctic phantom midges and ancient bunyavirid-like sequences in insect genomes". J Virol. 88 (16): 8783–94. doi:10.1128/JVI.00531-14. PMC 4136290. PMID 24850747.
- ↑ "Taxonomy browser (California encephalitis virus)". www.ncbi.nlm.nih.gov. Archived from the original on 16 October 2022. Retrieved 26 January 2023.
- 1 2 3 4 Abudurexiti, Abulikemu; Adkins, Scott; Alioto, Daniela; Alkhovsky, Sergey V.; Avšič-Županc, Tatjana; Ballinger, Matthew J.; Bente, Dennis A.; Beer, Martin; Bergeron, Éric; Blair, Carol D.; Briese, Thomas; Buchmeier, Michael J.; Burt, Felicity J.; Calisher, Charles H.; Cháng, Chénchén; Charrel, Rémi N.; Choi, Il Ryong; Clegg, J. Christopher S.; de la Torre, Juan Carlos; de Lamballerie, Xavier; Dèng, Fēi; Di Serio, Francesco; Digiaro, Michele; Drebot, Michael A.; Duàn, Xiǎoméi; Ebihara, Hideki; Elbeaino, Toufic; Ergünay, Koray; Fulhorst, Charles F.; Garrison, Aura R.; Gāo, George Fú; Gonzalez, Jean-Paul J.; Groschup, Martin H.; Günther, Stephan; Haenni, Anne-Lise; Hall, Roy A.; Hepojoki, Jussi; Hewson, Roger; Hú, Zhìhóng; Hughes, Holly R.; Jonson, Miranda Gilda; Junglen, Sandra; Klempa, Boris; Klingström, Jonas; Kòu, Chūn; Laenen, Lies; Lambert, Amy J.; Langevin, Stanley A.; Liu, Dan; Lukashevich, Igor S.; Luò, Tāo; Lǚ, Chuánwèi; Maes, Piet; de Souza, William Marciel; Marklewitz, Marco; Martelli, Giovanni P.; Matsuno, Keita; Mielke-Ehret, Nicole; Minutolo, Maria; Mirazimi, Ali; Moming, Abulimiti; Mühlbach, Hans-Peter; Naidu, Rayapati; Navarro, Beatriz; Nunes, Márcio Roberto Teixeira; Palacios, Gustavo; Papa, Anna; Pauvolid-Corrêa, Alex; Pawęska, Janusz T.; Qiáo, Jié; Radoshitzky, Sheli R.; Resende, Renato O.; Romanowski, Víctor; Sall, Amadou Alpha; Salvato, Maria S.; Sasaya, Takahide; Shěn, Shū; Shí, Xiǎohóng; Shirako, Yukio; Simmonds, Peter; Sironi, Manuela; Song, Jin-Won; Spengler, Jessica R.; Stenglein, Mark D.; Sū, Zhèngyuán; Sūn, Sùróng; Táng, Shuāng; Turina, Massimo; Wáng, Bó; Wáng, Chéng; Wáng, Huálín; Wáng, Jūn; Wèi, Tàiyún; Whitfield, Anna E.; Zerbini, F. Murilo; Zhāng, Jìngyuàn; Zhāng, Lěi; Zhāng, Yànfāng; Zhang, Yong-Zhen; Zhāng, Yújiāng; Zhou, Xueping; Zhū, Lìyǐng; Kuhn, Jens H. (July 2019). "Taxonomy of the order Bunyavirales: update 2019". Archives of Virology. 164 (7): 1949–1965. doi:10.1007/s00705-019-04253-6. Archived from the original on 14 August 2022. Retrieved 29 January 2023.
- ↑ Akram, Sami M.; Mangat, Rupinder; Huang, Ben (2022). "Hantavirus Cardiopulmonary Syndrome". StatPearls. StatPearls Publishing. Archived from the original on 29 August 2021. Retrieved 27 January 2023.
- 1 2 3 "Bunyavirales | Viral Hemorrhagic Fevers (VHFs) | CDC". www.cdc.gov. 3 September 2021. Archived from the original on 7 January 2023. Retrieved 29 January 2023.
- ↑ Horne, Kate McElroy; Vanlandingham, Dana L. (2014-11-13). "Bunyavirus-Vector Interactions". Viruses. 6 (11): 4373–4397. doi:10.3390/v6114373. ISSN 1999-4915. PMC 4246228. PMID 25402172.
- ↑ Lv Q, Zhang H, Tian L, Zhang R, Zhang Z, Li J, Tong Y, Fan H, Carr MJ, Shi W. Novel sub-lineages, recombinants and reassortants of severe fever with thrombocytopenia syndrome virus. Ticks Tick Borne Dis. 2017 Mar;8(3):385-390. doi: 10.1016/j.ttbdis.2016.12.015. Epub 2017 Jan 3. PMID 28117273
- ↑ He CQ, Ding NZ. Discovery of severe fever with thrombocytopenia syndrome bunyavirus strains originating from intragenic recombination. J Virol. 2012 Nov;86(22):12426-30. doi: 10.1128/JVI.01317-12. Epub 2012 Aug 29. PMID 22933273
- ↑ Patrick R. Murray, Ken S. Rosenthal and Michael A. Pfaller (2008-12-24). Medical Microbiology, 6e (6 ed.). Philadelphia: Mosby. ISBN 9780323054706.
- ↑ Bennett, John E.; Dolin, Raphael; Blaser, Martin J. (8 August 2019). Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. Elsevier Health Sciences. p. 2169. ISBN 978-0-323-55027-7. Archived from the original on 2 February 2023. Retrieved 28 January 2023.
- 1 2 3 Bayot, Marlon L.; King, Kevin C. (2022). "Biohazard Levels". StatPearls. StatPearls Publishing. Archived from the original on 2022-10-02. Retrieved 2023-01-28.
- ↑ "Crimean-Congo Hemorrhagic Fever (CCHF) | CDC". www.cdc.gov. 27 February 2019. Archived from the original on 21 December 2022. Retrieved 26 January 2023.
- ↑ Jonsson, Colleen B.; Figueiredo, Luiz Tadeu Moraes; Vapalahti, Olli (April 2010). "A Global Perspective on Hantavirus Ecology, Epidemiology, and Disease". Clinical Microbiology Reviews. 23 (2): 412–441. doi:10.1128/CMR.00062-09. Archived from the original on 8 December 2022. Retrieved 26 January 2023.
- ↑ Holden, Preston; Hess, A. D. (30 October 1959). "Cache Valley Virus, a Previously Undescribed Mosquito-Borne Agent". Science. 130 (3383): 1187–1188. doi:10.1126/science.130.3383.1187. Retrieved 26 January 2023.
- ↑ "La Crosse Virus Neuroinvasive Disease --- Missouri, 2009". www.cdc.gov. Archived from the original on 10 November 2022. Retrieved 26 January 2023.
- ↑ Shope, Robert E. (1996). "Bunyaviruses". Medical Microbiology (4th ed.). University of Texas Medical Branch at Galveston. ISBN 978-0-9631172-1-2. Archived from the original on 2022-08-10. Retrieved 26 January 2023.
- ↑ Lvov, D. K.; Alkhovsky, S. V. (30 December 2018). "Bunyavirales Order". Problems of Particularly Dangerous Infections (4): 15–19. doi:10.21055/0370-1069-2018-4-15-19. Retrieved 24 January 2023.
External links
- Viralzone: Bunyaviridae Archived 2010-06-13 at the Wayback Machine
- ICTVdb Index of Viruses—Bunyaviridae Archived 2009-10-04 at the Wayback Machine
- The Big Picture Book of Viruses: Bunyaviridae Archived 2016-05-11 at the Wayback Machine
- Bunyaviridae Genomes—database search results from the Viral Bioinformatics Resource Center
- Virus Pathogen Database and Analysis Resource (ViPR): Bunyaviridae Archived 2020-04-04 at the Wayback Machine
- "Bunyaviridae". NCBI Taxonomy Browser. 11571. Archived from the original on 2023-01-23. Retrieved 2023-01-08.