Teicoplanin
 | |
| Clinical data | |
|---|---|
| Pronunciation | /ˌtaɪkoʊˈpleɪnɪn/ TY-koh-PLAY-nin |
| Trade names | Targocid |
| AHFS/Drugs.com | International Drug Names |
| Pregnancy category |
|
| Routes of administration | Intravenous, intramuscular |
| ATC code | |
| Legal status | |
| Legal status | |
| Pharmacokinetic data | |
| Bioavailability | 90% (given IM) |
| Protein binding | 90% to 95% |
| Metabolism | Nil |
| Elimination half-life | 70 to 100 hours |
| Excretion | Kidney (97% unchanged) |
| Identifiers | |
IUPAC name
| |
| CAS Number | |
| PubChem CID | |
| DrugBank | |
| ChemSpider | |
| UNII | |
| KEGG | |
| ChEMBL | |
| NIAID ChemDB | |
| CompTox Dashboard (EPA) | |
| Chemical and physical data | |
| Formula | Variable |
| Molar mass | 1564.3 to 1907.7 g/mol |
| Melting point | 260 °C (500 °F) (dec.) |
InChI
| |
| | |
Teicoplanin is an antibiotic used in the prophylaxis and treatment of serious infections caused by Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus and Enterococcus faecalis. It is a semisynthetic glycopeptide antibiotic with a spectrum of activity similar to vancomycin. Its mechanism of action is to inhibit bacterial cell wall synthesis.[2]
Teicoplanin is marketed by Sanofi-Aventis under the trade name Targocid. Other trade names include Ticocin marketed by Cipla(India).
Oral teicoplanin has been demonstrated to be effective in the treatment of pseudomembranous colitis and Clostridium difficile-associated diarrhoea, with comparable efficacy with vancomycin.[3]
Its strength is considered to be due to the length of the hydrocarbon chain.[4]
Susceptibility data
Teicoplanin targets peptidoglycan synthesis making it an effective antimicrobial against Gram-positive bacteria including Staphylococci and Clostridium spp. The following represents MIC susceptibility data for a few medically significant pathogens:
- Clostridium difficile: 0.06 μg/ml - 0.5 μg/ml
- Staphylococcus aureus: ≤0.06 μg/ml - ≥128 μg/ml
- Staphylococcus epidermidis: ≤0.06 μg/ml - 32 μg/ml
Chemistry
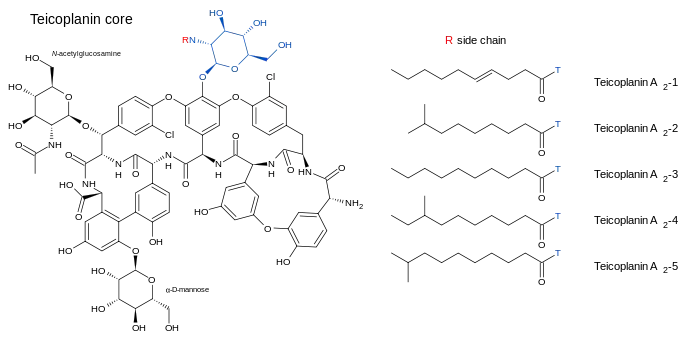
Teicoplanin (TARGOCID, marketed by Sanofi Aventis Ltd) is actually a mixture of several compounds, five major (named teicoplanin A2-1 through A2-5) and four minor (named teicoplanin RS-1 through RS-4).[6] All teicoplanins share a same glycopeptide core, termed teicoplanin A3-1 — a fused ring structure to which two carbohydrates (mannose and N-acetylglucosamine) are attached. The major and minor components also contain a third carbohydrate moiety — β-D-glucosamine — and differ only by the length and conformation of a side-chain attached to it.
The structures of the teicoplanin core and the side-chains that characterize the five major teicoplanin compounds are shown below.

Teicoplanin refers to a complex of related natural products isolated from the fermentation broth of a strain of Actinoplanes teichomyceticus,[7] consisting of a group of five structures. These structures possess a common aglycone, or core, consisting of seven amino acids bound by peptide and ether bonds to form a four-ring system. These five structures differ by the identity of the fatty acyl side-chain attached to the sugar. The origin of these seven amino acids in the biosynthesis of teicoplanin was studied by 1H and 13C nuclear magnetic resonance.[8] The studies indicate amino acids 4-Hpg, 3-Cl-Tyr, and 3-chloro-β-hydroxytyrosine are derived from tyrosine, and the amino acid 3,5-dihydroxyphenylglycine (3,5-Dpg) is derived from acetate. Teicoplanin contains 6 non-proteinogenic amino acids and three sugar moieties, N-acyl-β-D-glucosamine, N-acetyl-β-D-glucosamine, and D-mannose.
Gene cluster
The study of the genetic cluster encoding the biosynthesis of teicoplanin identified 49 putative open reading frames (ORFs) involved in the compound's biosynthesis, export, resistance, and regulation. Thirty-five of these ORFs are similar to those found in other glycopeptide gene clusters. The function of each of these genes is described by Li and co-workers.[9] A summary of the gene layout and purpose is shown below.
Gene layout. The genes are numbered. The letters L and R designate transcriptional direction. The presence of the * symbol means a gene is found after NRPs, which are represented by A, B, C, and D. Based on the figure from: Li, T-L.; Huang, F.; Haydock, S. F.; Mironenko, T.; Leadlay, P. F.; Spencer, J. B. Chemistry & Biology. 2004, 11, p. 109.
[11-L] [10-L] [9-R] [8-R] [7-R] [6-R] [5-R] [4-L][3-L] [2-L] [1-R] [A-R] [B-R] [C-R] [D-R] [1*-R] [2*-R] [3*-R] [4*-R] [5*-R] [6*-R] [7*-R] [8*-R] [9*-R] [10*-R] [11*-R] [12*-R] [13*-R] [14*-R] [15*-R] [16*-R] [17*-R] [18*-R] [19*-R] [20*-R] [21*-R] [22*-R] [23*-R] [24*-R] [25*-L] [26*-L] [27*-R] [28*-R] [29*-R] [30*-R][31*-R] [32*-L] [33*-L] [34*-R]
| Enzyme produced by gene sequence | Regulatory proteins | Other enzymes | Resistant enzymes | Β-hydroxy-tyrosine and 4-hydroxy-phenylglycin biosynthetic enzymes | Glycosyl transferases | Peptide synthetases | P450 oxygenases | Halogenase | 3,5-dihydroxy phenylglycin biosynthetic enzymes |
| Genes | 11, 10, 3, 2, 15*, 16*, 31* | 9, 8, 1*, 2*, 4*, 11*, 13*, 21*, 26*, 27*, 30*, 32*, 33*, 34* | 7, 6, 5 | 4, 12*, 14*, 22*, 23*, 24*, 25*, 28*, 29* | 1, 3*, 10* | A, B, C, D | 5*, 6*, 7*, 9* | 8* | 17*, 18*, 19*, 20*, 23* |
Heptapeptide backbone synthesis
The heptapeptide backbone of teicoplanin is assembled by the nonribosomal peptide synthetases (NRPSs) TeiA, TeiB, TeiC and TeiD. Together these comprise seven modules, each containing a number of domains, with each module responsible for the incorporation of a single amino acid. Modules 1, 4, and 5 activate L-4-Hpg as the aminoacyl-AMP, modules 2 and 6 activate L-Tyr, and modules 3 and 7 activate L-3,5-Dpg. The activated amino acids are covalently bound to the NRPS as thioesters by a phosphopantetheine cofactor, which is attached to the peptidyl carrier protein (PCP) domain. The enzyme bound amino acids are then joined by amide bonds by the action of the condensation (C) domain.
The heptapetide of teicoplanin contains 4 D-amino acids, formed by epimerization of the activated L-amino acids. Modules 2, 4 and 5 each contain an epimerization (E) domain which catalyzes this change. Module 1 does not contain an E domain, and epimerization is proposed to be catalysed by the C domain.[10] In all, six of the seven total amino acids of the teicoplanin backbone are composed of nonproteinogenic or modified amino acids. Eleven enzymes are coordinatively induced to produce these six required residues.[11] Teicoplanin contains two chlorinated positions, 2 (3-Cl-Tyr) and 6 (3-Cl-β-Hty). The halogenase Tei8* has been acts to catalyze the halogenation of both tyrosine residues. Chlorination occurs at the amino acyl-PCP level during the biosynthesis, prior to phenolic oxidative coupling, with the possibility of tyrosine or β-hydroxytyrosine being the substrate of chlorination.[12] Hydroxylation of the tyrosine residue of module 6 also occurs in trans during the assembly of the heptapeptide backbone.
Modification after heptapeptide backbone formation
Once the heptapeptide backbone has been formed, the linear enzyme-bound intermediate is cyclized.[11] Gene disruption studies indicate cytochrome P450 oxygenases as the enzymes that performs the coupling reactions. The X-domain in the final NRPS module is required to recruit the oxygenase enzymes.[13] OxyB forms the first ring by coupling residues 4 and 6, and OxyE then couples residues 1 and 3. OxyA couples residues 2 and 4, followed by the formation of a C-C bond between residues 5 and 7 by OxyC.[14] The regioselectivity and atropisomer selectivity of these probable one-electron coupling reactions has been suggested to be due to the folding and orientation requirements of the partially crossed-linked substrates in the enzyme active site.[11] The coupling reactions are shown below.
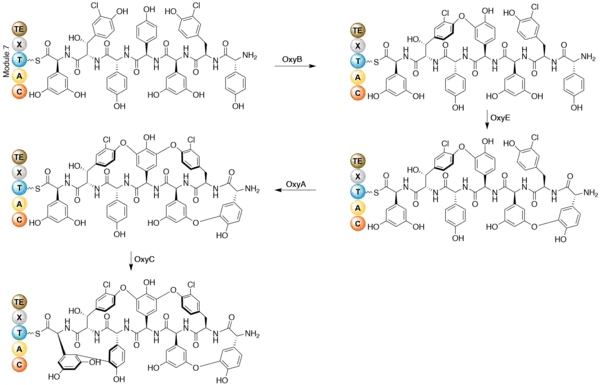
Specific glycosylation has been shown to occur after the formation of the heptpeptide aglycone.[15] Three separate glycosyl transferases are required for the glycosylation of the teicoplanin aglycone. Tei10* catalyses the addition of GlcNAc to residue 4, followed by deacetylation by Tei2*. The acyl chain (produced by the action of Tei30* and Tei13*) is then added by Tei11*. Tei1 then adds a second GlcNAc to the β-hydroxyl group of residue 6, followed by mannosylation of residue 7 catalysed by Tei3*.[16]
References
- ↑ https://www.ema.europa.eu/documents/psusa/teicoplanin-list-nationally-authorised-medicinal-products-emea/h/n/psr/s/0025_en.pdf
- ↑ Reynolds PE (November 1989). "Structure, biochemistry and mechanism of action of glycopeptide antibiotics". European Journal of Clinical Microbiology & Infectious Diseases. 8 (11): 943–50. doi:10.1007/BF01967563. PMID 2532132. S2CID 21551939.
- ↑ de Lalla F, Nicolin R, Rinaldi E, Scarpellini P, Rigoli R, Manfrin V, Tramarin A (October 1992). "Prospective study of oral teicoplanin versus oral vancomycin for therapy of pseudomembranous colitis and Clostridium difficile-associated diarrhea". Antimicrobial Agents and Chemotherapy. 36 (10): 2192–6. doi:10.1128/AAC.36.10.2192. PMC 245474. PMID 1444298.
- ↑ Gilpin M, Milner P (1997). "Resisting changes -- Over the past 40 years the glycopeptide antibiotics have played a crucial role in treating bacterial infections. But how long can it continue ?". Royal Society of Chemistry. Archived from the original on 2002-12-21. Retrieved 2006-10-15. - includes picture of Teicoplanin's structure.
- ↑ Teicoplanin Susceptibility and Minimum Inhibitory Concentration (MIC) Data
- ↑ Bernareggi A, Borghi A, Borgonovi M, Cavenaghi L, Ferrari P, Vékey K, et al. (August 1992). "Teicoplanin metabolism in humans". Antimicrobial Agents and Chemotherapy. 36 (8): 1744–9. doi:10.1128/AAC.36.8.1744. PMC 192040. PMID 1416858.
- ↑ Jung HM, Jeya M, Kim SY, Moon HJ, Kumar Singh R, Zhang YW, Lee JK (September 2009). "Biosynthesis, biotechnological production, and application of teicoplanin: current state and perspectives". Applied Microbiology and Biotechnology. 84 (3): 417–28. doi:10.1007/s00253-009-2107-4. PMID 19609520. S2CID 45038487.
- ↑ Heydorn A, Petersen BO, Duus JO, Bergmann S, Suhr-Jessen T, Nielsen J (March 2000). "Biosynthetic studies of the glycopeptide teicoplanin by (1)H and (13)C NMR". The Journal of Biological Chemistry. 275 (9): 6201–6. doi:10.1074/jbc.275.9.6201. PMID 10692413.
- ↑ Li TL, Huang F, Haydock SF, Mironenko T, Leadlay PF, Spencer JB (January 2004). "Biosynthetic gene cluster of the glycopeptide antibiotic teicoplanin: characterization of two glycosyltransferases and the key acyltransferase". Chemistry & Biology. 11 (1): 107–19. doi:10.1016/j.chembiol.2004.01.001. PMID 15113000.
- ↑ Kaniusaite M, Tailhades J, Kittilä T, Fage CD, Goode RJ, Schittenhelm RB, Cryle MJ (May 2020). "Understanding the early stages of peptide formation during the biosynthesis of teicoplanin and related glycopeptide antibiotics". The FEBS Journal. 288 (2): 507–529. doi:10.1111/febs.15350. PMID 32359003. S2CID 218481859.
- 1 2 3 Kahne D, Leimkuhler C, Lu W, Walsh C (February 2005). "Glycopeptide and lipoglycopeptide antibiotics". Chemical Reviews. 105 (2): 425–48. doi:10.1021/cr030103a. PMID 15700951.
- ↑ Kittilä T, Kittel C, Tailhades J, Butz D, Schoppet M, Büttner A, et al. (September 2017). "Halogenation of glycopeptide antibiotics occurs at the amino acid level during non-ribosomal peptide synthesis". Chemical Science. 8 (9): 5992–6004. doi:10.1039/C7SC00460E. PMC 5620994. PMID 28989629.
- ↑ Haslinger K, Peschke M, Brieke C, Maximowitsch E, Cryle MJ (May 2015). "X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis". Nature. 521 (7550): 105–9. Bibcode:2015Natur.521..105H. doi:10.1038/nature14141. PMID 25686610. S2CID 4466657.
- ↑ Peschke M, Brieke C, Cryle MJ (October 2016). "F-O-G Ring Formation in Glycopeptide Antibiotic Biosynthesis is Catalysed by OxyE". Scientific Reports. 6 (1): 35584. Bibcode:2016NatSR...635584P. doi:10.1038/srep35584. PMC 5067714. PMID 27752135.
- ↑ Kaplan J, Korty BD, Axelsen PH, Loll PJ (May 2001). "The role of sugar residues in molecular recognition by vancomycin". Journal of Medicinal Chemistry. 44 (11): 1837–40. doi:10.1021/jm0005306. PMID 11356118.
- ↑ Yushchuk O, Ostash B, Pham TH, Luzhetskyy A, Fedorenko V, Truman AW, Horbal L (August 2016). "Characterization of the Post-Assembly Line Tailoring Processes in Teicoplanin Biosynthesis". ACS Chemical Biology. 11 (8): 2254–64. doi:10.1021/acschembio.6b00018. PMID 27285718.