Rhodopsin-like receptors
Rhodopsin-like receptors are a family of proteins that comprise the largest group of G protein-coupled receptors.[2]
| Rhodopsin-like receptors | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
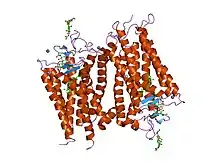 Structure of rhodopsin: A G protein-coupled receptor.[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | 7tm_1 | ||||||||||
| Pfam | PF00001 | ||||||||||
| Pfam clan | GPCR_A | ||||||||||
| InterPro | IPR000276 | ||||||||||
| PROSITE | PDOC00211 | ||||||||||
| SCOP2 | 1f88 / SCOPe / SUPFAM | ||||||||||
| OPM superfamily | 6 | ||||||||||
| OPM protein | 1gzm | ||||||||||
| CDD | cd00637 | ||||||||||
| |||||||||||
Scope
G-protein-coupled receptors, GPCRs, constitute a vast protein family that encompasses a wide range of functions (including various autocrine, paracrine, and endocrine processes). They show considerable diversity at the sequence level, on the basis of which they can be separated into distinct groups. GPCRs are usually described as "superfamily" because they embrace a group of families for which there are indications of evolutionary relationship, but between which there is no statistically significant similarity in sequence.[2] The currently known superfamily members include the rhodopsin-like GPCRs (this family), the secretin-like GPCRs, the cAMP receptors, the fungal mating pheromone receptors, and the metabotropic glutamate receptor family. There is a specialised database for GPCRs.[3]
Function
The rhodopsin-like GPCRs themselves represent a widespread protein family that includes hormone, neuropeptide, neurotransmitter, and light receptors, all of which transduce extracellular signals through interaction with guanine nucleotide-binding (G) proteins. Although their activating ligands vary widely in structure and character, the amino acid sequences of the receptors are very similar and are believed to adopt a common structural framework comprising 7 transmembrane (TM) helices.[4][5][6]
Classes
Rhodopsin-like GPCRs have been classified into the following 19 subgroups (A1-A19) based on a phylogenetic analysis.[7]
Subfamily A1
- Chemokine receptor InterPro: IPR000355
- Chemokine (C-C motif) receptor 1 (CCR1, CKR1)
- Chemokine (C-C motif) receptor 2 (CCR2, CKR2)
- Chemokine (C-C motif) receptor 3 (CCR3, CKR3)
- Chemokine (C-C motif) receptor 4 (CCR4, CKR4)
- Chemokine (C-C motif) receptor 5 (CCR5, CKR5)
- Chemokine (C-C motif) receptor 8 (CCR8, CKR8)
- Chemokine (C-C motif) receptor-like 2 (CCRL2, CKRX)
- chemokine (C motif) receptor 1 (XCR1, CXC1) InterPro: IPR005393
- chemokine (C-X3-C motif) receptor 1 (CX3CR1, C3X1) InterPro: IPR005387
- GPR137B (GPR137B, TM7SF1)
Subfamily A2
- Chemokine receptor InterPro: IPR000355
- Chemokine (C-C motif) receptor-like 1 (CCRL1 CCRL1, CCR11)
- Chemokine (C-C motif) receptor 6 (CCR6, CKR6)
- Chemokine (C-C motif) receptor 7 (CCR7, CKR7)
- Chemokine (C-C motif) receptor 9 (CCR9, CKR9)
- Chemokine (C-C motif) receptor 10 (CCR10, CKRA)
- CXC chemokine receptors InterPro: IPR001053
- Interleukin-8 InterPro: IPR000174 (IL8R)
- Adrenomedullin receptor (GPR182)
- Duffy blood group, chemokine receptor (DARC, DUFF)
- G Protein-coupled Receptor 30 (GPER, CML2, GPCR estrogen receptor)
Subfamily A3
- Angiotensin II receptor InterPro: IPR000248
- Angiotensin II receptor, type 1 (AGTR1, AG2S)
- Angiotensin II receptor, type 2 (AGTR2, AG22)
- Apelin receptor (AGTRL1, APJ) InterPro: IPR003904
- Bradykinin receptor InterPro: IPR000496
- Bradykinin receptor B1 (BDKRB1, BRB1)
- Bradykinin receptor B2 (BDKRB2, BRB2)
- GPR15 (GPR15, GPRF)
- GPR25 (GPR25)
Subfamily A4
- Opioid receptor InterPro: IPR001418
- delta Opioid receptor (OPRD1, OPRD)
- kappa Opioid receptor (OPRK1, OPRK)
- mu Opioid receptor (OPRM1, OPRM)
- Nociceptin receptor (OPRL1, OPRX)
- Somatostatin receptor InterPro: IPR000586
- Somatostatin receptor 1 (SSTR1, SSR1)
- Somatostatin receptor 2 (SSTR2, SSR2)
- Somatostatin receptor 3 (SSTR3, SSR3)
- Somatostatin receptor 4 (SSTR4, SSR4)
- Somatostatin receptor 5 (SSTR5, SSR5)
- GPCR neuropeptide receptor InterPro: IPR009150
- Neuropeptides B/W receptor 1 (NPBWR1, GPR7)
- Neuropeptides B/W receptor 2 (NPBWR2, GPR8)
- GPR1 orphan receptor (GPR1) InterPro: IPR002275
Subfamily A5
- Galanin receptor InterPro: IPR000405
- Galanin receptor 1 (GALR1, GALR)
- Galanin receptor 2 (GALR2, GALS)
- Galanin receptor 3 (GALR3, GALT)
- Cysteinyl leukotriene receptor InterPro: IPR004071
- Leukotriene B4 receptor InterPro: IPR003981
- Relaxin receptor InterPro: IPR008112
- Relaxin/insulin-like family peptide receptor 1 (RXFP1, LGR7)
- Relaxin/insulin-like family peptide receptor 2 (RXFP2, GPR106)
- Relaxin/insulin-like family peptide receptor 3 (RXFP3, SALPR)
- Relaxin/insulin-like family peptide receptor 4 (RXFP4, GPR100/GPR142)
- KiSS1-derived peptide receptor (GPR54) (KISS1R) InterPro: IPR008103
- Melanin-concentrating hormone receptor 1 (MCHR1, GPRO) InterPro: IPR008361
- Urotensin-II receptor (UTS2R, UR2R) InterPro: IPR000670
Subfamily A6
- Cholecystokinin receptor InterPro: IPR009126
- Cholecystokinin A receptor (CCKAR, CCKR)
- Cholecystokinin B receptor (CCKBR, GASR)
- Neuropeptide FF receptor InterPro: IPR005395
- Neuropeptide FF receptor 1 (NPFFR1, FF1R)
- Neuropeptide FF receptor 2 (NPFFR2, FF2R)
- Orexin receptor InterPro: IPR000204
- Hypocretin (orexin) receptor 1 (HCRTR1, OX1R)
- Hypocretin (orexin) receptor 2 (HCRTR2, OX2R)
- Vasopressin receptor InterPro: IPR001817
- Gonadotropin releasing hormone receptor (GNRHR, GRHR) InterPro: IPR001658
- Pyroglutamylated RFamide peptide receptor (QRFPR, GPR103)
- GPR22 (GPR22, GPRM)
- GPR176 (GPR176, GPR)
Subfamily A7
- Bombesin receptor InterPro: IPR001556
- Endothelin receptor InterPro: IPR000499
- Endothelin receptor type A (EDNRA, ET1R)
- Endothelin receptor type B (EDNRB, ETBR)
- GPR37 (GPR37, ETBR-LP2) InterPro: IPR003909
- Neuromedin U receptor InterPro: IPR005390
- Neurotensin receptor InterPro: IPR003984
- Neurotensin receptor 1 (NTSR1, NTR1)
- Neurotensin receptor 2 (NTSR2, NTR2)
- Thyrotropin-releasing hormone receptor (TRHR, TRFR) InterPro: IPR009144
- Growth hormone secretagogue receptor (GHSR) InterPro: IPR003905
- GPR39 (GPR39)
- Motilin receptor (MLNR, GPR38)
Subfamily A8
- Anaphylatoxin receptors InterPro: IPR002234
- C3a receptor (C3AR1, C3AR)
- C5a receptor (C5AR1, C5AR)
- Chemokine-like receptor 1 (CMKLR1, CML1) InterPro: IPR002258
- Formyl peptide receptor InterPro: IPR000826
- Formyl peptide receptor 1 (FPR1, FMLR)
- Formyl peptide receptor-like 1 (FPRL1, FML2)
- Formyl peptide receptor-like 2 (FPRL2, FML1)
- MAS1 oncogene InterPro: IPR000820
- GPR1 (GPR1)
- GPR32 (GPR32, GPRW)
- GPR44 (GPR44)
- GPR77 (GPR77, C5L2)
Subfamily A9
- Melatonin receptor InterPro: IPR000025
- Melatonin receptor 1A (MTNR1A, ML1A)
- Melatonin receptor 1B (MTNR1B, ML1B)
- Neurokinin receptor InterPro: IPR001681
- Tachykinin receptor 1 (TACR1, NK1R)
- Tachykinin receptor 2 (TACR2, NK2R)
- Tachykinin receptor 3 (TACR3, NK3R)
- Neuropeptide Y receptor InterPro: IPR000611
- Neuropeptide Y receptor Y1 (NPY1R, NY1R)
- Neuropeptide Y receptor Y2 (NPY2R, NY2R)
- Pancreatic polypeptide receptor 1 (PPYR1, NY4R)
- Neuropeptide Y receptor Y5 (NPY5R, NY5R)
- Prolactin-releasing peptide receptor (PRLHR, GPRA) InterPro: IPR001402
- Prokineticin receptor 1 (PROKR1, GPR73)
- Prokineticin receptor 2 (PROKR2, PKR2)
- GPR19 (GPR19, GPRJ)
- GPR50 (GPR50, ML1X)
- GPR75 (GPR75)
- GPR83 (GPR83, GPR72)
Subfamily A10
- Glycoprotein hormone receptor InterPro: IPR002131
- Leucine-rich repeat-containing G protein-coupled receptor 4 (LGR4, GPR48)
- Leucine-rich repeat-containing G protein-coupled receptor 5 (LGR5, GPR49)
- Leucine-rich repeat-containing G protein-coupled receptor 6 (LGR6)
Subfamily A11
- GPR40-related receptor InterPro: IPR013312
- Free fatty acid receptor 1 (FFAR1, GPR40)
- Free fatty acid receptor 2 (FFAR2, GPR43)
- Free fatty acid receptor 3 (FFAR3, GPR41)
- GPR42 (GPR42, FFAR1L)
- P2 purinoceptor InterPro: IPR002286
- Hydroxycarboxylic acid receptor 1 (HCAR1, GPR81)
- Hydroxycarboxylic acid receptor 2, Niacin receptor 1 (HCAR2, GPR109A)
- Hydroxycarboxylic acid receptor 3, Niacin receptor 2 (HCAR3, GPR109B, HM74)
- GPR31 (GPR31, GPRV)
- GPR82 (GPR82)
- Oxoglutarate (alpha-ketoglutarate) receptor 1 (OXGR1, GPR80)
- Succinate receptor 1 (SUCNR1, GPR91)
Subfamily A12
- P2 purinoceptor InterPro: IPR002286
- Purinergic receptor P2Y12 (P2RY12)
- Purinergic receptor P2Y13 (P2RY13, GPR86) InterPro: IPR008109
- Purinergic receptor P2Y14 (P2RY14, UDP-glucose receptor, KI01) InterPro: IPR005466
- GPR34 (GPR34)
- GPR87 (GPR87)
- GPR171 (GPR171, H963)
- Platelet-activating factor receptor (PTAFR, PAFR) InterPro: IPR002282
Subfamily A13
- Cannabinoid receptor InterPro: IPR002230
- Cannabinoid receptor 1 (brain) (CNR1, CB1R)
- Cannabinoid receptor 2 (macrophage) (CNR2, CB2R)
- Lysophosphatidic acid receptor InterPro: IPR004065
- Sphingosine 1-phosphate receptor InterPro: IPR004061
- Melanocortin/ACTH receptor InterPro: IPR001671
- Melanocortin 1 receptor (MC1R, MSHR)
- Melanocortin 3 receptor (MC3R)
- Melanocortin 4 receptor (MC4R)
- Melanocortin 5 receptor (MC5R)
- ACTH receptor (MC2R), ACTR)
- GPR3 (GPR3)
- GPR6 (GPR6)
- GPR12 (GPR12, GPRC)
Subfamily A14
- Eicosanoid receptor InterPro: IPR008365
- Prostaglandin D2 receptor (PTGDR, PD2R)
- Prostaglandin E1 receptor (PTGER1, PE21)
- Prostaglandin E2 receptor (PTGER2, PE22)
- Prostaglandin E3 receptor (PTGER3, PE23)
- Prostaglandin E4 receptor (PTGER4, PE24)
- Prostaglandin F receptor (PTGFR, PF2R)
- Prostaglandin I2 (prostacyclin) receptor (PTGIR, PI2R)
- Thromboxane A2 receptor (TBXA2R, TA2R)
Subfamily A15
- Lysophosphatidic acid receptor InterPro: IPR004065
- P2 purinoceptor InterPro: IPR002286
- Purinergic receptor P2Y10 (P2RY10, P2Y10)
- Protease-activated receptor InterPro: IPR003912
- Epstein-Barr virus induced gene 2 (lymphocyte-specific G protein-coupled receptor) (GPR183)
- Proton-sensing G protein-coupled receptors
- GPR17 (GPR17, GPRH)
- GPR18 (GPR18, GPRI)
- GPR20 (GPR20, GPRK)
- GPR35 (GPR35)
- GPR55 (GPR55)
- Coagulation factor II receptor (F2R, THRR)
Subfamily A16
- Opsins InterPro: IPR001760[8]
- Rhodopsin (RHO, OPSD)
- Opsin 1 (cone pigments), short-wave-sensitive (color blindness, tritan) (OPN1SW, OPSB) (blue-sensitive opsin)
- Opsin 1 (cone pigments), medium-wave-sensitive (color blindness, deutan) (OPN1MW, OPSG) (green-sensitive opsin)
- Opsin 1 (cone pigments), long-wave-sensitive (color blindness, protan) (OPN1LW, OPSR) (red-sensitive opsin)
- Opsin 3, Panopsin (OPN3)
- Opsin 4, Melanopsin (OPN4)
- Opsin 5 (OPN5, GPR136)
- Retinal G protein coupled receptor (RGR)
- Retinal pigment epithelium-derived rhodopsin homolog (RRH, OPSX) (visual pigment-like receptor opsin) InterPro: IPR001793
Subfamily A17
Subfamily A18
- Histamine H1 receptor (HRH1, HH1R) InterPro: IPR000921
- Histamine H3 receptor (HRH3) InterPro: IPR003980
- Histamine H4 receptor (HRH4) InterPro: IPR008102
- Adenosine receptor InterPro: IPR001634
- Muscarinic acetylcholine receptor InterPro: IPR000995
- GPR21 (GPR21, GPRL)
- GPR27 (GPR27)
- GPR45 (GPR45, PSP24)
- GPR52 (GPR52)
- GPR61 (GPR61)
- GPR62 (GPR62)
- GPR63 (GPR63)
- GPR78 (GPR78)
- GPR84 (GPR84)
- GPR85 (GPR85)
- GPR88 (GPR88)
- GPR101 (GPR101)
- GPR161 (GPR161, RE2)
- GPR173 (GPR173, SREB3)
Subfamily A19
References
- Palczewski K, Kumasaka T, Hori T, et al. (August 2000). "Crystal structure of rhodopsin: A G protein-coupled receptor". Science. 289 (5480): 739–45. Bibcode:2000Sci...289..739P. doi:10.1126/science.289.5480.739. PMID 10926528.
- Attwood TK, Findlay JB (1994). "Fingerprinting G-protein-coupled receptors". Protein Eng. 7 (2): 195–203. doi:10.1093/protein/7.2.195. PMID 8170923.
- "Information system for G protein-coupled receptors". GPCRDB. www.gpcr.org. Archived from the original on 2009-04-22. Retrieved 2008-12-05.
- Birnbaumer L (1990). "G proteins in signal transduction". Annu. Rev. Pharmacol. Toxicol. 30: 675–705. doi:10.1146/annurev.pa.30.040190.003331. PMID 2111655.
- Gilman AG, Casey PJ (1988). "G protein involvement in receptor-effector coupling". J. Biol. Chem. 263 (6): 2577–2580. PMID 2830256.
- Attwood TK, Findlay JB (1993). "Design of a discriminating fingerprint for G-protein-coupled receptors". Protein Eng. 6 (2): 167–176. doi:10.1093/protein/6.2.167. PMID 8386361.
- Joost P, Methner A (2002). "Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands". Genome Biol. 3 (11): research0063.1–0063.16. doi:10.1186/gb-2002-3-11-research0063. PMC 133447. PMID 12429062.
- Terakita A (2005). "The opsins". Genome Biol. 6 (3): 213. doi:10.1186/gb-2005-6-3-213. PMC 1088937. PMID 15774036.
- Nordström KJ, Sällman Almén M, Edstam MM, Fredriksson R, Schiöth HB (September 2011). "Independent HHsearch, Needleman—Wunsch-based, and motif analyses reveal the overall hierarchy for most of the G protein-coupled receptor families". Molecular Biology and Evolution. 28 (9): 2471–80. doi:10.1093/molbev/msr061. PMID 21402729.
External links
- Vriend G, Horn F, Oliveira L, Bywater RP, Cohen FE. "GPCRDB (G Protein-Coupled Receptor Data Base): sequence-derived data". Archived from the original on 2007-11-14. Retrieved 2007-12-10.
- Horn F, Bettler E, Oliveira L, Campagne F, Cohen FE, Vriend G (2003). "GPCRDB information system for G protein-coupled receptors". Nucleic Acids Res. 31 (1): 294–7. doi:10.1093/nar/gkg103. PMC 165550. PMID 12520006. This database includes multiple sequence alignments of all GPCR families and sub-families.