Small nucleolar RNA SNORA73
In molecular biology, the small nucleolar RNA SNORA73 (also called U17/E1 RNA) belongs to the H/ACA class of small nucleolar RNAs (snoRNAs). Vertebrate U17 is intron-encoded and ranges in length from 200-230 nucleotides, longer than most snoRNAs. It is one of the most abundant snoRNAs in human cells and is essential for the cleavage of pre-rRNA within the 5' external transcribed spacer (ETS).[1] This cleavage leads to the formation of 18S rRNA. Regions of the U17 RNA are complementary to rRNA and act as guides for RNA/RNA interactions, although these regions do not seem to be well conserved between organisms.[2]
| Small nucleolar RNA SNORA73 family | |
|---|---|
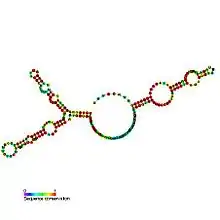 Predicted secondary structure and sequence conservation of SNORA73 | |
| Identifiers | |
| Symbol | SNORA73 |
| Alt. Symbols | U17 |
| Rfam | RF00045 |
| Other data | |
| RNA type | Gene; snRNA; snoRNA; HACA-box |
| Domain(s) | Eukaryota |
| GO | GO:0006396 GO:0005730 |
| SO | SO:0000594 |
| PDB structures | PDBe |
There is evidence that SNORA73 (isoforms: SNORA73A and SNORA73B) functions as a regulator of chromatin function.[3] SNORA73 is chromatin-associated RNA (caRNA) and stably linked to chromatin.[4] Notably, SNORA73 can bind to PARP1, leading to the activation of its ADPRylation (PAR) function.[5] SNORA73 Interacts with the PARP1 DNA-Binding Domain. In addition, the snoRNA-activated PARP1 ADPRylates DDX21 in cells to promote cell proliferation.[6]
See also
References
- Enright CA, Maxwell ES, Eliceiri GL, Sollner-Webb B (November 1996). "5'ETS rRNA processing facilitated by four small RNAs: U14, E3, U17, and U3". RNA. 2 (11): 1094–9. PMC 1369439. PMID 8903340.
- Cervelli M, Cecconi F, Giorgi M, Annesi F, Oliverio M, Mariottini P (February 2002). "Comparative structure analysis of vertebrate U17 small nucleolar RNA (snoRNA)". Journal of Molecular Evolution. 54 (2): 166–79. Bibcode:2002JMolE..54..166C. doi:10.1007/s00239-001-0065-2. PMID 11821910. S2CID 9424738.
- Schubert, T; Pusch, MC; Diermeier, S; Benes, V; Kremmer, E; Imhof, A; Längst, G (9 November 2012). "Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin". Molecular Cell. 48 (3): 434–44. doi:10.1016/j.molcel.2012.08.021. PMID 23022379.
- Schubert, T; Pusch, MC; Diermeier, S; Benes, V; Kremmer, E; Imhof, A; Längst, G (9 November 2012). "Df31 protein and snoRNAs maintain accessible higher-order structures of chromatin". Molecular Cell. 48 (3): 434–44. doi:10.1016/j.molcel.2012.08.021. PMID 23022379.
- Kim, DS; Camacho, CV; Nagari, A; Malladi, VS; Challa, S; Kraus, WL (19 September 2019). "Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21". Molecular Cell. 75 (6): 1270–1285.e14. doi:10.1016/j.molcel.2019.06.020. PMC 6754283. PMID 31351877.
- Kim, DS; Camacho, CV; Nagari, A; Malladi, VS; Challa, S; Kraus, WL (19 September 2019). "Activation of PARP-1 by snoRNAs Controls Ribosome Biogenesis and Cell Growth via the RNA Helicase DDX21". Molecular Cell. 75 (6): 1270–1285.e14. doi:10.1016/j.molcel.2019.06.020. PMC 6754283. PMID 31351877.