Small nucleolar RNA snR69
In molecular biology, snoRNA snR69 is a non-coding RNA (ncRNA) molecule which functions in the modification of other small nuclear RNAs (snRNAs). This type of modifying RNA is usually located in the nucleolus of the eukaryotic cell which is a major site of snRNA biogenesis. It is known as a small nucleolar RNA (snoRNA) and also often referred to as a guide RNA. snoRNA snR69 belongs to the C/D box class of snoRNAs which contain the conserved sequence motifs known as the C box (UGAUGA) and the D box (CUGA). Most of the members of the box C/D family function in directing site-specific 2'-O-methylation of substrate RNAs.[1] snoRNA snR69 was initially discovered using a computational screen of the Saccharomyces cerevisiae genome.[2]
| Small nucleolar RNA snR69 | |
|---|---|
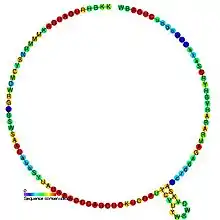 Predicted secondary structure and sequence conservation of snosnR69 | |
| Identifiers | |
| Symbol | snosnR69 |
| Rfam | RF00475 |
| Other data | |
| RNA type | Gene; snRNA; snoRNA; CD-box |
| Domain(s) | Eukaryota |
| GO | GO:0005730 GO:0006396 |
| SO | SO:0000593 |
| PDB structures | PDBe |
References
- Galardi S, Fatica A, Bachi A, Scaloni A, Presutti C, Bozzoni I (October 2002). "Purified box C/D snoRNPs are able to reproduce site-specific 2'-O-methylation of target RNA in vitro". Molecular and Cellular Biology. 22 (19): 6663–8. doi:10.1128/MCB.22.19.6663-6668.2002. PMC 134041. PMID 12215523.
- Lowe TM, Eddy SR (February 1999). "A computational screen for methylation guide snoRNAs in yeast". Science. 283 (5405): 1168–71. Bibcode:1999Sci...283.1168L. doi:10.1126/science.283.5405.1168. PMID 10024243.
External links
This article is issued from Wikipedia. The text is licensed under Creative Commons - Attribution - Sharealike. Additional terms may apply for the media files.