PZM21
 | |
 | |
| Identifiers | |
|---|---|
IUPAC name
| |
| CAS Number | |
| PubChem CID | |
| DrugBank | |
| ChemSpider | |
| Chemical and physical data | |
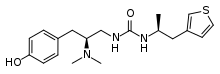
| Formula | C19H27N3O2S |
| Molar mass | 361.50 g·mol−1 |
| 3D model (JSmol) | |
SMILES
| |
InChI
| |
PZM21 is an experimental opioid analgesic drug that is being researched for the treatment of pain.[1] It is claimed to be a functionally selective μ-opioid receptor agonist which produces μ-opioid receptor mediated G protein signaling, with potency and efficacy similar to morphine, but with less β-arrestin 2 recruitment.[2] However, recent reports highlight that this might be due to its low intrinsic efficacy,[3] rather than functional selectivity or 'G protein bias' as initially reported. In tests on mice, PZM21 was slightly less potent than morphine or TRV130 as an analgesic, but also had significantly reduced adverse effects, with less constipation than morphine, and very little respiratory depression, even at high doses.[4] This research was described as a compelling example of how modern high-throughput screening techniques can be used to discover new chemotypes with specific activity profiles, even at targets such as the μ-opioid receptor which have already been thoroughly investigated.[5] More recent research has suggested however that at higher doses, PZM21 is capable of producing classic opioid side effects such as respiratory depression and development of tolerance and may have only limited functional selectivity.[6]
See also
References
- ↑ Kostic M (September 2016). "Biasing Opioid Receptors and Cholesterol as a Player in Developmental Biology". Cell Chemical Biology. 23 (9): 1039–1040. doi:10.1016/j.chembiol.2016.09.007. PMID 27662248.
- ↑ Manglik A, Lin H, Aryal DK, McCorvy JD, Dengler D, Corder G, et al. (September 2016). "Structure-based discovery of opioid analgesics with reduced side effects". Nature. 537 (7619): 185–190. Bibcode:2016Natur.537..185M. doi:10.1038/nature19112. PMC 5161585. PMID 27533032.
- ↑ Gillis A, Gondin AB, Kliewer A, Sanchez J, Lim HD, Alamein C, et al. (March 2020). "Low intrinsic efficacy for G protein activation can explain the improved side effect profiles of new opioid agonists". Science Signaling. 13 (625): eaaz3140. doi:10.1126/scisignal.aaz3140. PMID 32234959. S2CID 214771721.
- ↑ Manglik A, Lin H, Aryal DK, McCorvy JD, Dengler D, Corder G, et al. (September 2016). "Structure-based discovery of opioid analgesics with reduced side effects". Nature. 537 (7619): 185–190. Bibcode:2016Natur.537..185M. doi:10.1038/nature19112. PMC 5161585. PMID 27533032.
- ↑ Kieffer BL (September 2016). "Drug discovery: Designing the ideal opioid". Nature. 537 (7619): 170–171. Bibcode:2016Natur.537..170K. doi:10.1038/nature19424. PMID 27533037.
- ↑ Hill R, Disney A, Conibear A, Sutcliffe K, Dewey W, Husbands S, et al. (July 2018). "The novel μ-opioid receptor agonist PZM21 depresses respiration and induces tolerance to antinociception". British Journal of Pharmacology. 175 (13): 2653–2661. doi:10.1111/bph.14224. PMC 6003631. PMID 29582414.